Transient Expression Vector Construction, Subcellular Localisation, and Evaluation of Antiviral Potential of Flagellin BP8-2
Abstract
:1. Introduction
2. Results
2.1. Amplification and Identification of BP8-2 Gene
2.2. Construction and Sequencing of Recombinant Plasmid
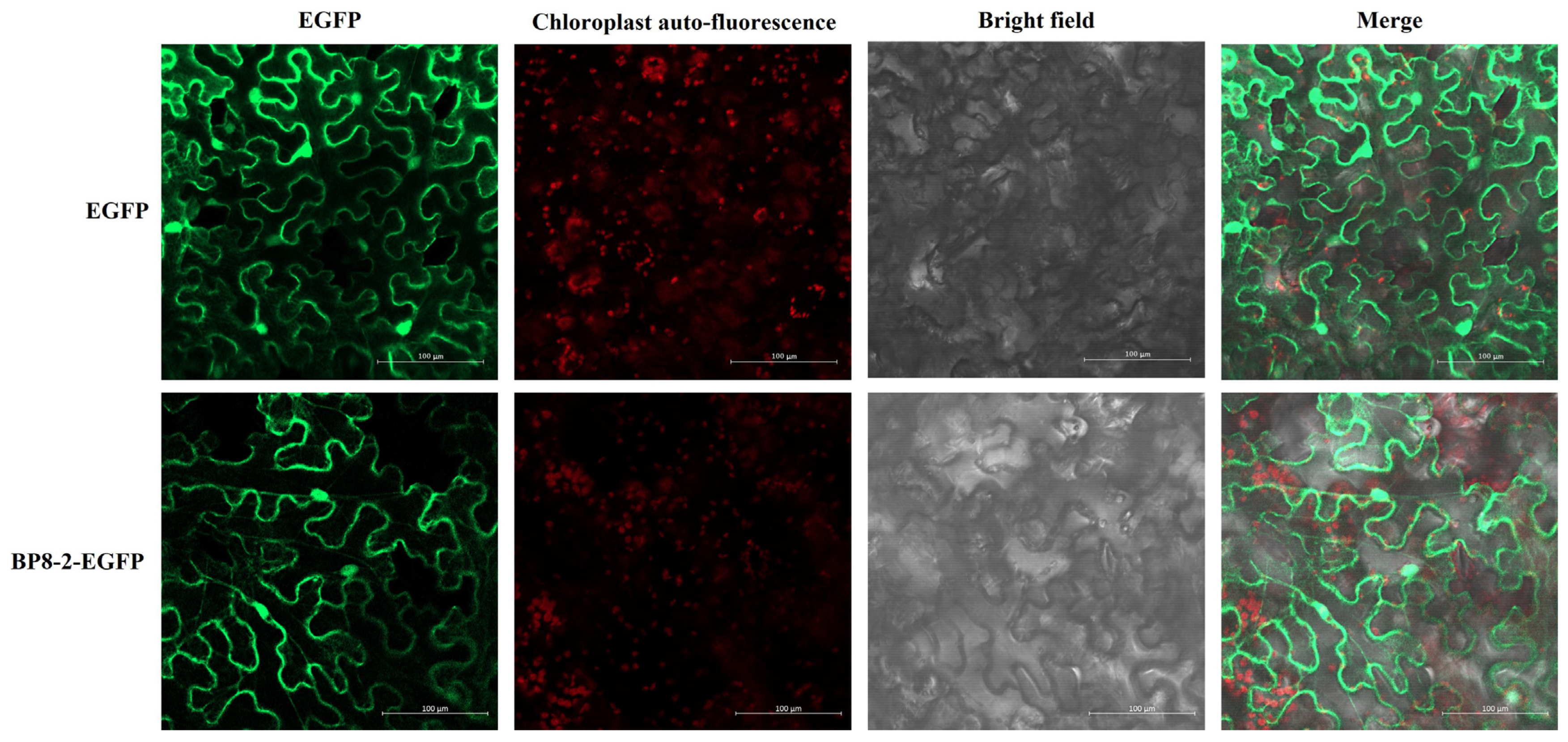
2.3. Subcellular Localisation and Transient Expression of BP8-2
2.4. Purification of BP8-2 Proteins
2.5. Anti-TMV Activities of BP8-2
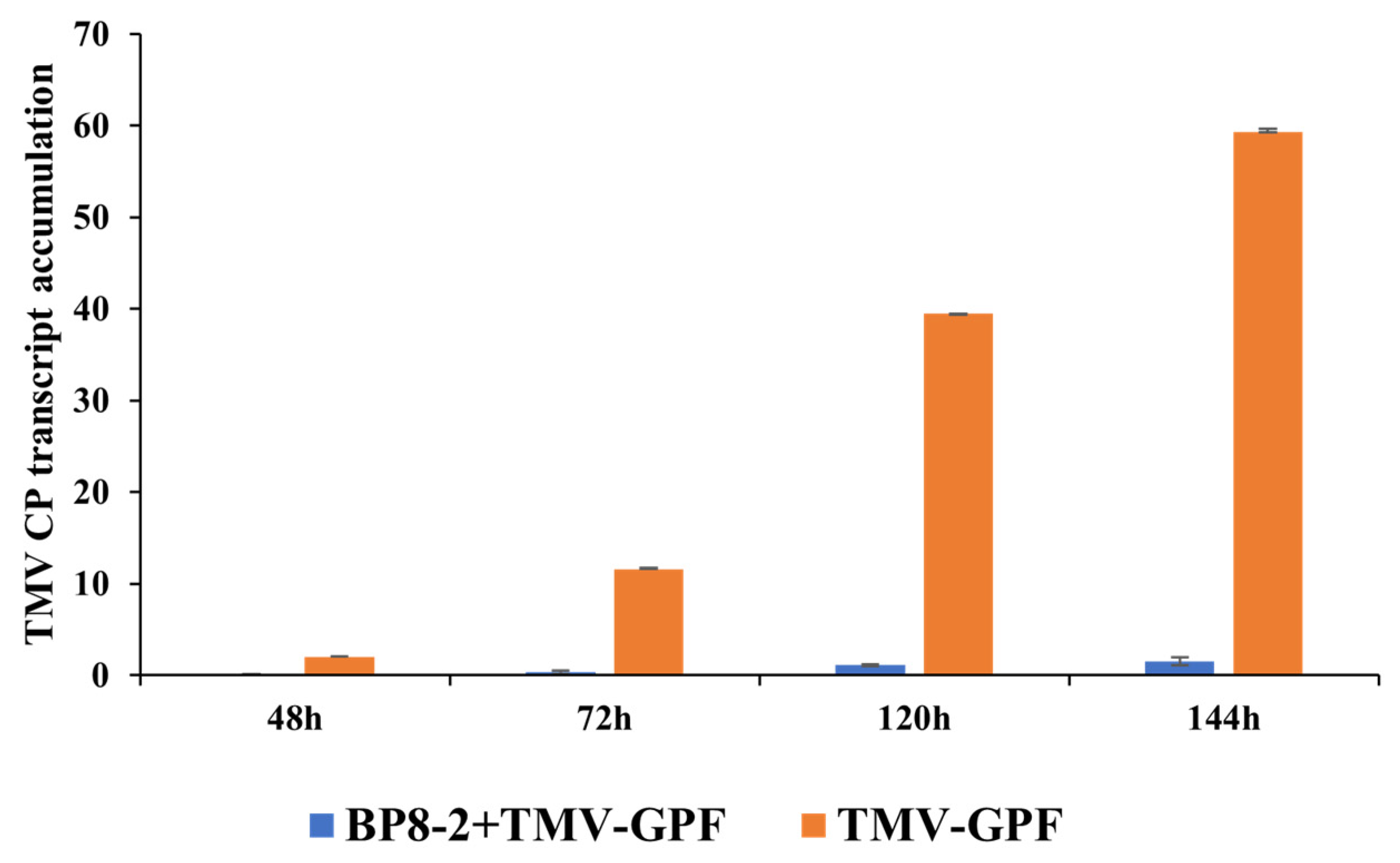
2.6. Effects of BP8-2 on TMV Infection Movement
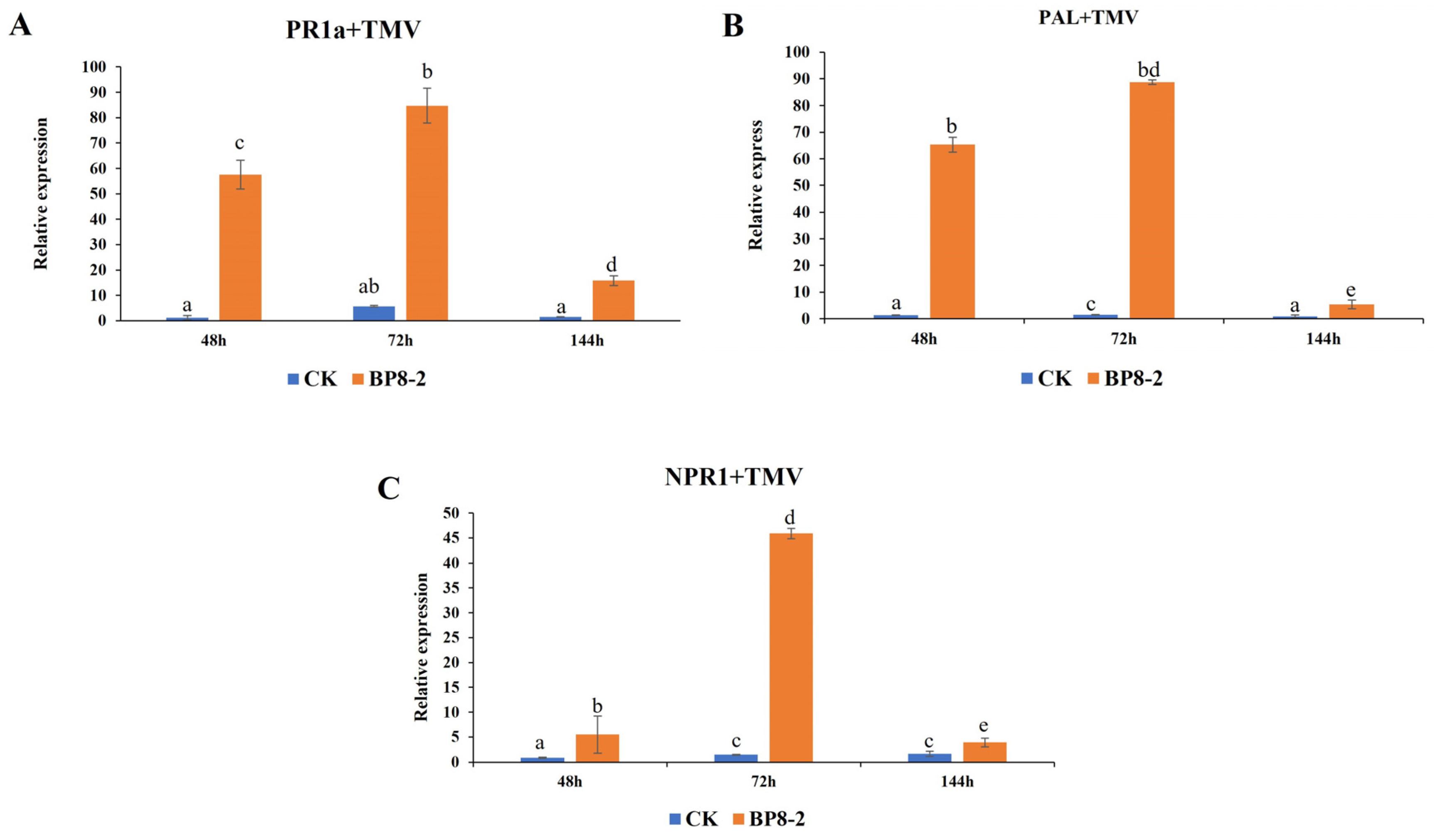
2.7. Expression of Defence-Related Genes
3. Discussion
4. Material and Methods
4.1. Chemicals and Materials
4.2. Virus Purification
4.3. Cloning of BP8-2 Gene
4.4. Construction and Sequencing of Recombinant Plasmid
4.5. Subcellular Localisation and Transient Expression of BP8-2
4.6. Purification of BP8-2 Protein by SDS-PAGE Electrophoresis
4.7. Inhibitory Effect of BP8-2 on TMV Infection
4.8. Protective Effect of BP8-2 on TMV Infection
4.9. Curative Effect of BP8-2 on TMV Infection
4.10. Effects of BP8-2 on TMV Infection Movement
4.11. Detection of the Expression of Defence-Related Genes by Real-Time Quantitative PCR
4.12. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, F.D.; Feng, G.H.; Chen, K.S. Burdock fructooligosaccharide induces resistance to tobacco mosaic virus in tobacco seedlings. Physiol. Mol. Plant Pathol. 2009, 74, 34–40. [Google Scholar] [CrossRef]
- Martin, W.H. The microbial diversity of inland waters. Curr. Opin. Biotechnol. 2006, 7, 256–261. [Google Scholar]
- Zhao, L.; Feng, C.; Wu, K.; Chen, W.; Chen, Y.; Hao, X.; Wu, Y. Advances and prospects in biogenic substances against plant virus: A review. Pestic. Biochem. Physiol. 2015, 135, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Erjaee, Z.; Shekarforoush, S.S.; Hosseinzadeh, S.; Dehghani, A.; Winter, D. Identification of antifungal intracellular proteins of endophytic Bacillus pumilus by LC–MS/MS analysis. Int. J. Pept. Res. Ther. 2020, 26, 2429–2435. [Google Scholar] [CrossRef]
- Zhou, W.W.; Zhang, X.L.; Zhang, B.; Wang, F.; Liang, Z.H.; Niu, T.G. Isolation and characterization of ZH14 with antiviral activity against tobacco mosaic virus. Can. J. Microbiol. 2008, 54, 441–449. [Google Scholar] [CrossRef]
- Shen, L.L.; Wang, F.L.; Yang, J.G.; Qian, Y.M.; Dong, X.W.; Zhan, H.X. Control of tobacco mosaic virus by Pseudomonas fluorescens CZ powder in greenhouses and the field. Crop. Prot. 2014, 56, 87–90. [Google Scholar] [CrossRef]
- Ju, Y.P.; Si, Y.Y.; Young, C.K.; Jin-Cheol, K.; Quang, L.D.; Jeong, J.K.; In, S.K. Antiviral peptide from Pseudomonas chlororaphis O6 against tobacco mosaic virus (TMV). J. Korean Soc. Appl. Biol. 2012, 55, 89–94. [Google Scholar]
- Thapa, S.P.; Lee, H.J.; Park, D.H.; Kim, S.K.; Cho, J.M.; Cho, S.; Hur, J.H.; Lim, C.K. Antiviral effects of the culture filtrate from Serratia marcescens Gsm01, against cucumber mosaic virus (CMV). Plant Pathol. J. 2009, 25, 369–375. [Google Scholar] [CrossRef]
- Ramos, H.C.; Rumbo, M.; Sirard, J.C. Bacterial flagellins: Mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004, 12, 509–517. [Google Scholar] [CrossRef]
- Altegoer, F.; Mukherjee, S.; Steinchen, W.; Bedrunka, P.; Linne, U.; Kearns, D.B.; Bange, G. FliS/flagellin/FliW heterotrimer couples type III secretion and flagellin homeostasis. Sci. Rep. 2018, 8, 11552. [Google Scholar] [CrossRef] [PubMed]
- Il Kim, M.; Lee, C.; Park, J.; Jeon, B.Y.; Hong, M. Crystal structure of Bacillus cereus flagellin and structure-guided fusion-protein designs. Sci. Rep. 2018, 8, 5814. [Google Scholar] [CrossRef]
- Qiu, D.W. Research progress of microbial protein pesticides. Chin. J. Biol. Control 2004, 20, 91–94. [Google Scholar]
- Li, Y.; Yao, H.; Liu, S.; Song, D.; Wu, C.; Zhang, S.; Gao, Q.; Zhang, L. The role of flagellin F in Vibrio parahaemolyticus-induced intestinal immunity and functional domain identification. Int. J. Biol. Macromol. 2021, 244, 125404. [Google Scholar] [CrossRef]
- Han, Q.; Wu, F.; Wang, X.; Hong, Q.; Liang, S.; Ren, A.; Liu, Q.; Zhao, M.; Tang, C. The bacterial lipopeptide iturins induce Verticillium dahliae cell death by affecting fungal signaling pathways and mediate plant defense responses involved in PAMP-triggered immunity. Environ. Microbiol. 2015, 17, 1166–1188. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Chen, Y.; Li, L.; Ma, Y.; Tong, Z.; Guo, D.; Sun, P.; An, D. Analysis and evaluation of the flagellin activity of Bacillus amyloliquefaciens Ba168 antimicrobial proteins against Penicillium expansum. Molecules 2022, 27, 4259. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.P.; Chakravarthy, S.; André CVelásquez Mclane, H.L.; Martin, G.B. Methods to study pamp-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant-Microbe Interact. 2010, 23, 991–999. [Google Scholar] [CrossRef]
- Xiang, T.T.; Ma, S.M.; Wo, E. Research progress of bacterial flagellin as a vaccine adjuvant. Chin. J. Bioloficals 2023, 36, 764–768. [Google Scholar]
- Leuzinger, K.; Dent, M.; Hurtado, J.; Jake, S. Efficient agroinfiltration of plants for high-level transient expression of recombinant proteins. J. Vis. Exp. JoVE 2013, 77, 50521. [Google Scholar]
- Wydro, M.; Kozubek, E.; Lehmann, P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 2006, 53, 289–298. [Google Scholar] [CrossRef]
- Hitzeroth, I.I.; Van Zyl, A.R. Transient expression of viral proteins in plants using Agrobacterium tumefaciens. In Vaccine Design. Methods in Molecular Biology; Thomas, S., Ed.; Humana: New York, NY, USA, 2016; pp. 581–595. [Google Scholar]
- Bidarigh Fard, A.; Nayeri, F.D.; Anbuhi, M.H. Transient expression of etanercept therapeutic proteinin tobacco (Nicotiana tabacum L.). Int. J. Biol. Macromol. 2019, 130, 483–490. [Google Scholar] [CrossRef]
- Zhang, J.X.; He, C.F.; Ye, X.G.; Dong, Y.M. Advances on Agrobacterium mediated transformation of Monocotyledons. Biotechnol. Bull. 2007, 2, 23–26. [Google Scholar]
- Li, X.J.; Wang, S.M.; Xie, Y.L.; He, M. Optimization of transformation conditions of tobacco by agrobacterium-mediated vacuum infiltration method. Jiangsu Agric. Sci. 2014, 42, 45–47. [Google Scholar]
- Zuo, Q.; Duan, Y.X.; Chen, L.J.; Zhu, X.F.; Wang, Y.Y. Application of protein subcellular localization method in plant pathology study. In Proceedings of the 2011 Annual Conference of Chinese Society of Plant Pathology, Yichang, China, 18–24 August 2011; pp. 435–439. [Google Scholar]
- Jiang, Y.Z.; He, L.H.; Liu, J.J. The methods and technologies for protein function study. Biotechnol. Bull. 2009, 9, 38–43. [Google Scholar]
- Chen, Y.H.; Ru, B.L.; Zhai, Y.Y.; Li, J.; Cheng, J.L. Screening and inhibitory effects of plant extracts against Tobacco mosaic virus (TMV). J. Plant Prot. 2018, 45, 463–469. [Google Scholar]
- Chen, Y.H.; Guo, D.S.; Lu, M.H.; Yue, J.Y.; Liu, Y.; Shang, C.M.; An, D.R.; Zhao, M.M. Inhibitory effect of osthole from Cnidium monnieri on Tobacco mosaic virus (TMV) Infection in Nicotiana glutinosa. Molecules 2019, 25, 65. [Google Scholar] [CrossRef]
- Mumford, R.; Macarthur, R.; Boonham, N. The role and challenges of new diagnostic technology in plant biosecurity. Food Secur. 2016, 8, 103–109. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of plant viruses and disease management: Relevance of genetic diversity and evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Elisabeth, K.; Dennis, J.L. Tobacco mosaic virus, not just a single component virus anymore. Mol. Plant Pathol. 2001, 2, 117–123. [Google Scholar]
- Verhage, L. How tobacco mosaic virus goes the distance. Plant J. 2021, 106, 894–895. [Google Scholar] [CrossRef]
- Ren, Y.; Armstrong, M.; Qi, Y.; McLellan, H.; Zhong, C.; Du, B.; Birch, P.R.; Tian, Z. Phytophthora infestans RXLR effectors target parallel steps in an immune signal transduction pathway. Plant Physiol. 2019, 180, 2227–2239. [Google Scholar] [CrossRef]
- Bai, S.; Liu, J.; Chang, C.; Zhang, L.; Maekawa, T.; Wang, Q.; Xiao, W.; Liu, Y.; Chai, J.; Takken, F.L.; et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012, 8, e1002752. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Berg, J.; Govers, F.; Bouwmeester, K. Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR 1. New Phytol. 2015, 207, 735–747. [Google Scholar] [CrossRef]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Bauer, Z.; Boller, T. Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in arabidopsis. Plant Cell 2001, 13, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Bittel, P.; Chinchilla, D.; Köchner, P.; Felix, G.; Shiu, S.H.; Boller, T. Molecular identification and characterization of the tomato flagel in receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristicaly differentperception specificities. Plant Mol. Biol. 2007, 64, 539–547. [Google Scholar] [CrossRef]
- Qiu, D.; Dong, Y.; Zhang, Y.; Li, S.; Shi, F. Plant immunity inducer development and application. Mol. Plant Microbe Interact. 2017, 30, 355. [Google Scholar]
- Wu, X.L.; Chen, H.M.; Wu, M.S. Activation of bacterial flagellin on plant immune defense response and its signaling mechanism. Plant Prot. 2011, 37, 12–16. [Google Scholar]
- Gooding, G.V.; Hebert, T.A. A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 1967, 57, 1285–1289. [Google Scholar] [PubMed]



| Drug | Inhibition Effect (%) | Protection Effect (%) | Curative Effect (%) |
|---|---|---|---|
| 100 μg/mL BP8-2 | 75.91 ± 5.33 a | 77.45 ± 5.17 a | 68.15 ± 5.93 a |
| 1000 times 8%Ningnanmycin SL | 63.92 ± 6.13 aA | 65.20 ± 2.02 aA | 61.67 ± 3.98 aA |
| Gene Name | Forward Primer Sequences (5′-3′) | Reverse Primer Sequences (5′-3′) |
|---|---|---|
| Actin | CTTGAAACAGCAAAGACCAGC | CTTGAAACAGCAAAGACCAGC |
| TMV CP | GACCTGACAAAAATGGAGAAGATCT | GAAAGCGGACAGAAACCCGCTG |
| Gene Name | Forward Primer Sequences (5′-3′) | Reverse Primer Sequences (5′-3′) |
|---|---|---|
| PR1a | GATGCCCATAACACAGCTCG | GATGCCCATAACACAGCTCG |
| PAL | ATTGCTGGTTTGCTCACTGG | TCCTTAGGCTGCAACTCGAA |
| NPR1 | GATACACGGTGCTGCATGTT | AAGCCTAGTGAGCCTCTTGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhong, J.; Lu, M.; Yang, C. Transient Expression Vector Construction, Subcellular Localisation, and Evaluation of Antiviral Potential of Flagellin BP8-2. Molecules 2024, 29, 3876. https://doi.org/10.3390/molecules29163876
Chen Y, Zhong J, Lu M, Yang C. Transient Expression Vector Construction, Subcellular Localisation, and Evaluation of Antiviral Potential of Flagellin BP8-2. Molecules. 2024; 29(16):3876. https://doi.org/10.3390/molecules29163876
Chicago/Turabian StyleChen, Yahan, Jianxin Zhong, Meihuan Lu, and Chengde Yang. 2024. "Transient Expression Vector Construction, Subcellular Localisation, and Evaluation of Antiviral Potential of Flagellin BP8-2" Molecules 29, no. 16: 3876. https://doi.org/10.3390/molecules29163876





