Study on the Rapid Limit Test for Six Sulfonamide Residues in Food Based on the TLC-SERS Method
Abstract
:1. Introduction
2. Results
2.1. Determination of the Characterization and Stability of Active Substrates
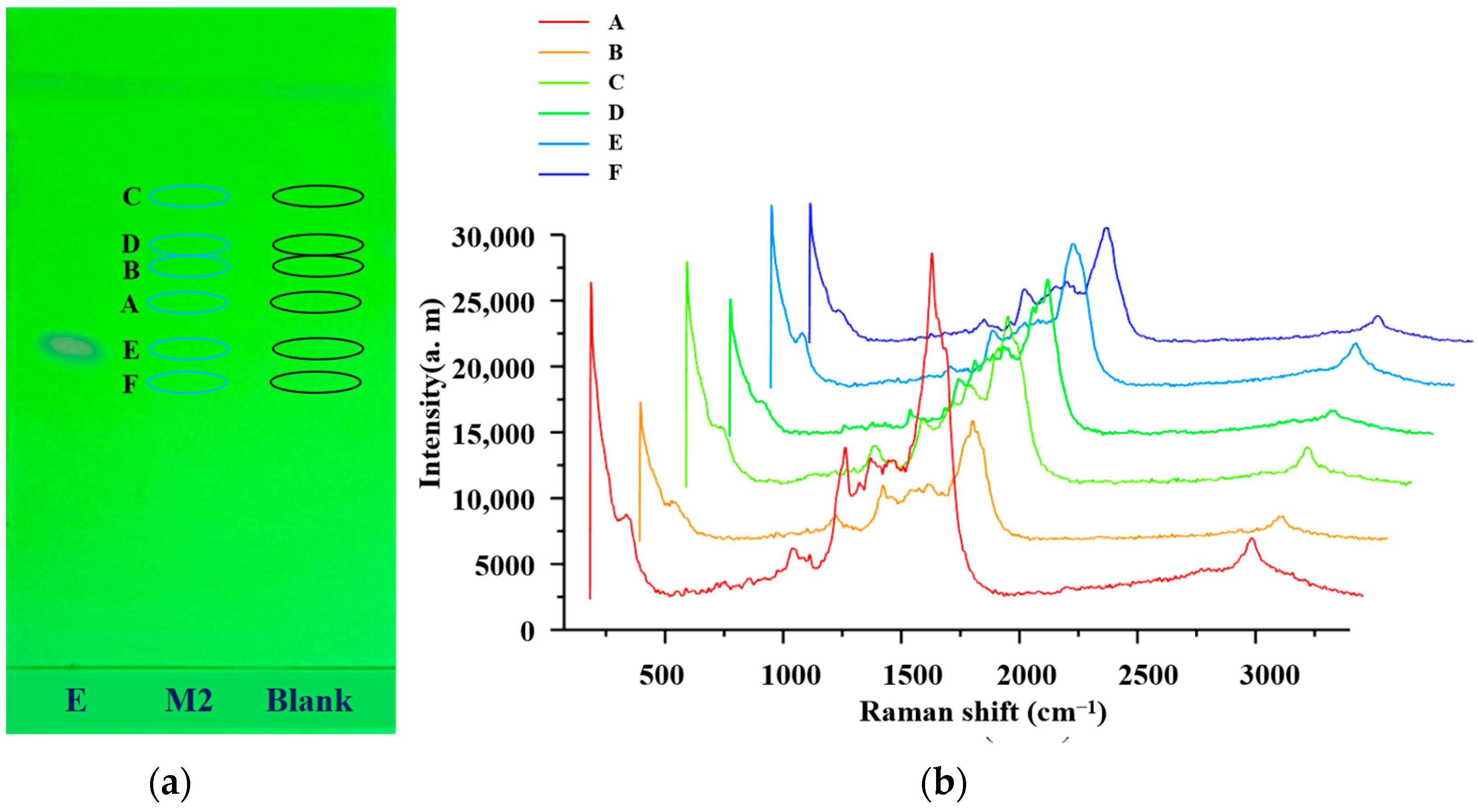
2.2. Relative Rf and Raman
2.3. In Situ SERS Detection of Sulfonamides
2.4. Calculation of EFs
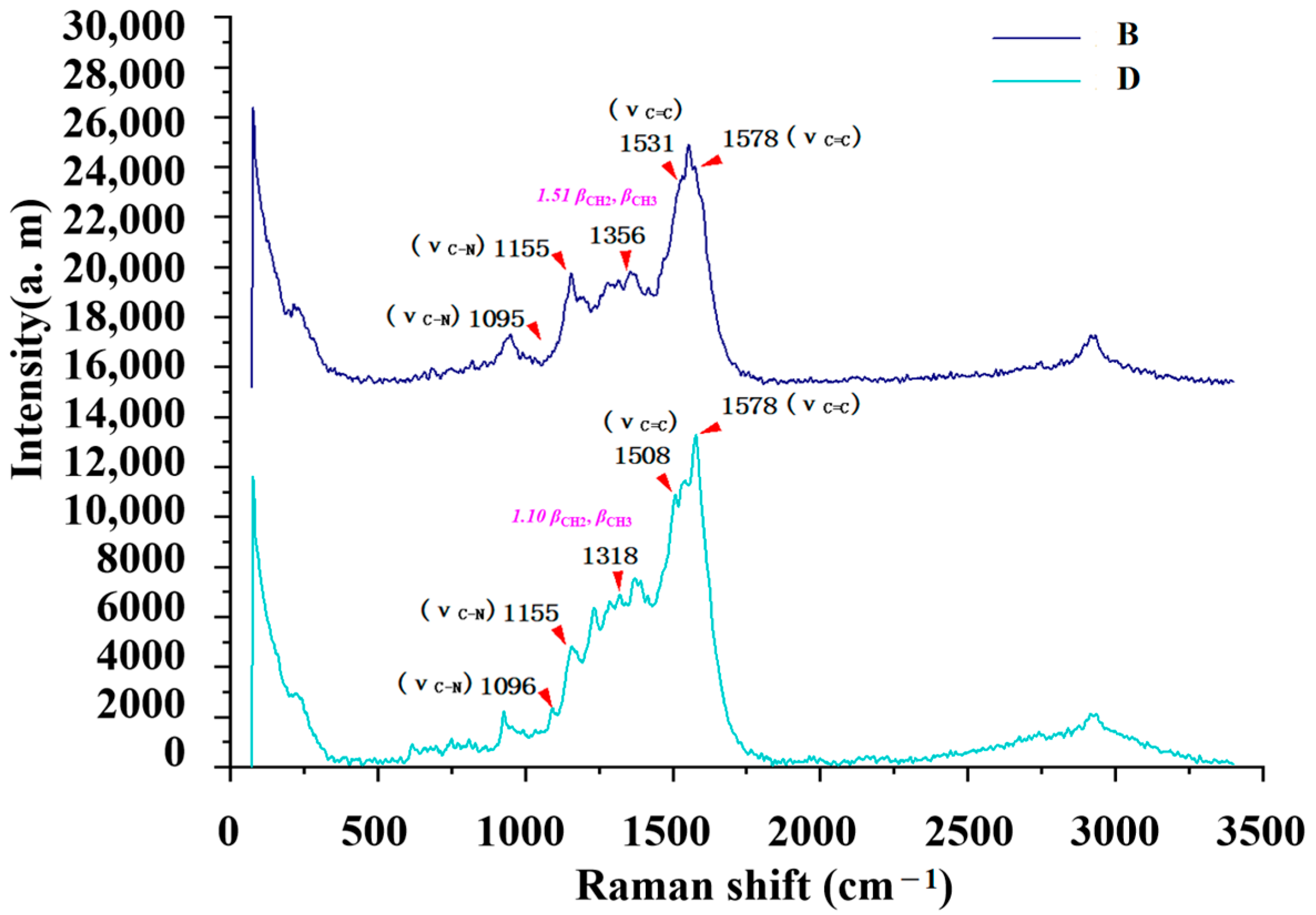
2.5. Comparative Analysis of the Raman and SERS Results
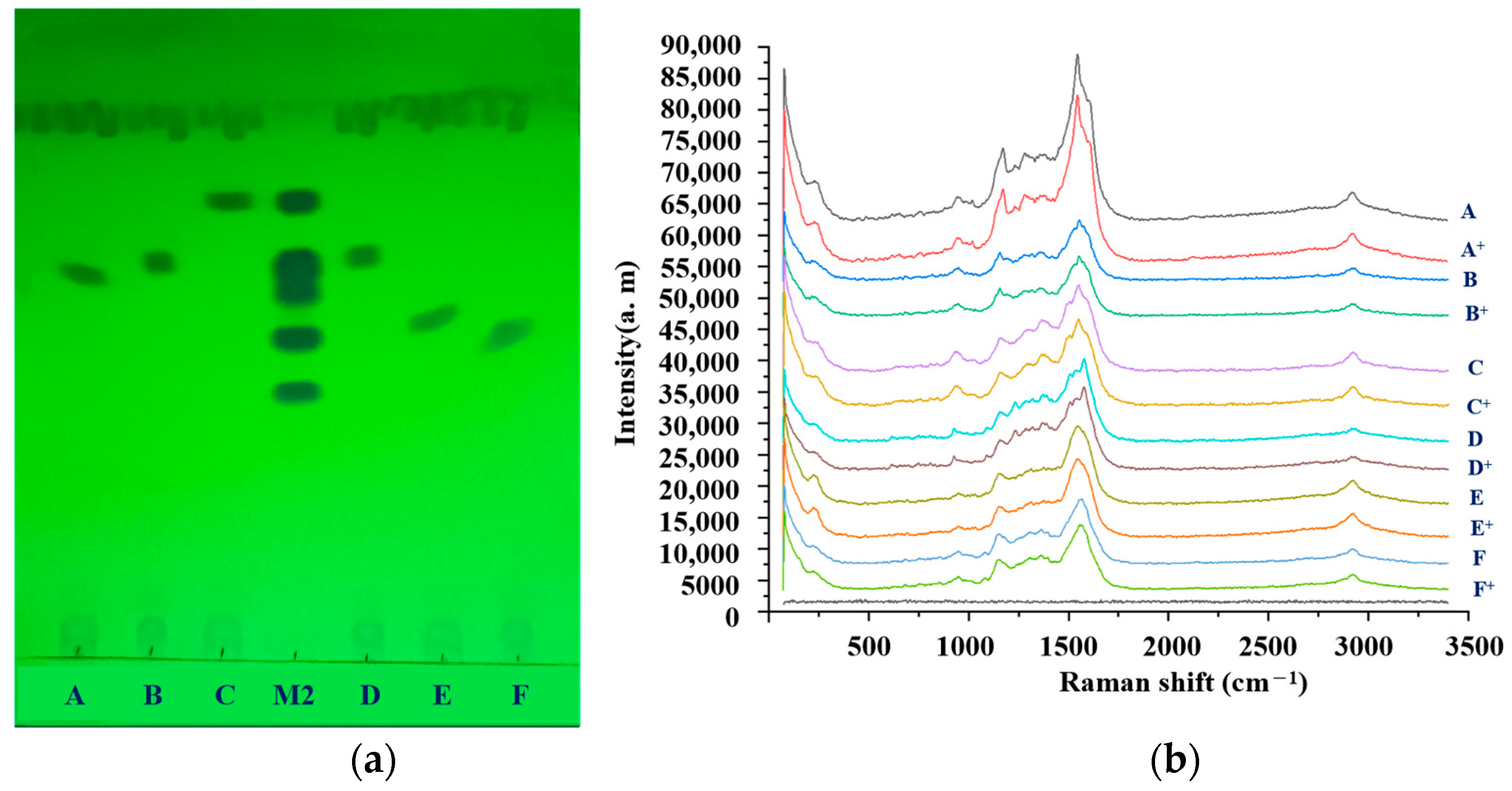
2.6. Identification by SERS Combined with Relative Rf
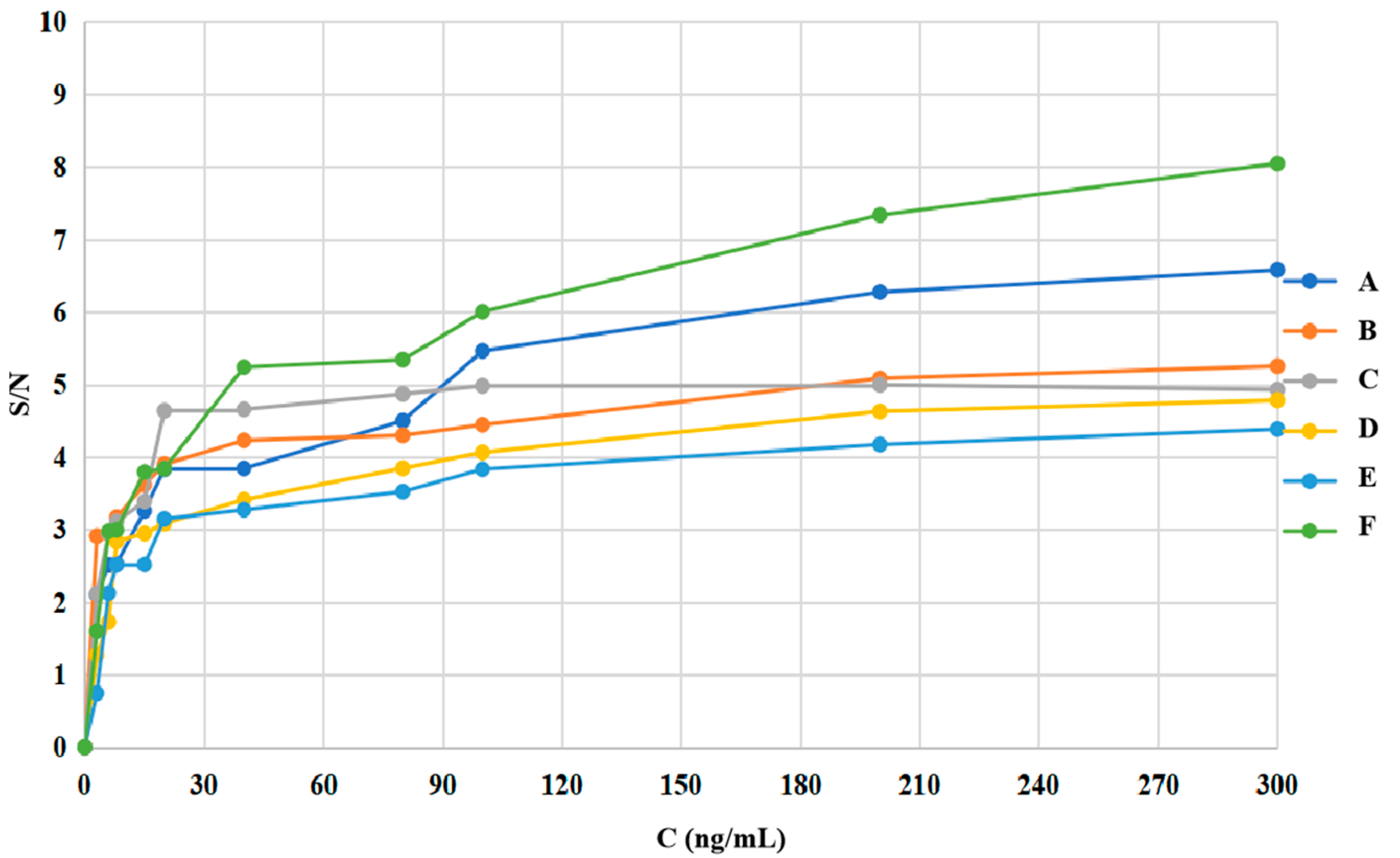
2.7. LOD Test
2.8. Stability Assessment and Specificity Testing
2.9. Simulated Positive Test
2.10. Limited Quantity Inspection Test of Real Samples
2.11. Comparison of the Results of TLC-SERS and UPLC-MS Detection
3. Materials and Methods
3.1. Materials
3.2. Apparatus and Conditions
3.3. Preparation of Silver Nanoparticles
3.4. Preparation of Reference and Mixed Reference Solutions
3.5. Preparation of Sample Solutions
3.6. TLC Test
3.7. TLC-SERS
3.8. UPLC-MS/MS Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Azevedo-Barbosa, H.; Dias, D.F.; Franco, L.L.; Hawkes, J.A.; Carvalho, D.T. From Antibacterial to antitumour agents: A brief review on the chemical and medicinal aspects of sulfonamides. Mini Rev. Med. Chem. 2020, 20, 2052–2066. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Arias, B.; Araya-Zúñiga, A.; Waterhouse-Garbanzo, J.; Rojas-Barrantes, Z.; Arguedas-Chacón, S.; Zavaleta-Monestel, E. Comprehensive review of sulfonamide hypersensitivity: Implications for clinical practice. Clin. Rev. Allergy Immunol. 2023, 65, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.R.; Paranhos, A.G.O.; de Aquino, S.F.; Silva, S.Q. Distribution of genetic elements associated with antibiotic resistance in treated and untreated animal husbandry waste and wastewater. Environ. Sci. Pollut. Res. Int. 2021, 28, 26380–26403. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Liu, X.; Dong, Y.; Yang, J.; Zhang, J.; He, S.; Yang, F.; Wang, Z.; Dong, Y. A Green HPLC method for determination of nine sulfonamides in milk and beef, and its greenness assessment with analytical eco-scale and greenness profile. J. AOAC Int. 2020, 103, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.M.; Habib, N.M.; Emam, A.A.; Mahmoud, H.M.; Abdelwhab, N.S. Chromatographic determination of sulfasalazine and its active metabolites: Greenness assessment and application to spiked human plasma. Biomed. Chromatogr. 2020, 34, e4804. [Google Scholar] [CrossRef] [PubMed]
- Queral-Beltran, A.; Marín-García, M.; Lacorte, S.; Tauler, R. UV-Vis absorption spectrophotometry and LC-DAD-MS-ESI(+)-ESI(-) coupled to chemometrics analysis of the monitoring of sulfamethoxazole degradation by chlorination, photodegradation, and chlorination/photodegradation. Anal. Chim. Acta 2023, 1276, 341563. [Google Scholar] [CrossRef] [PubMed]
- Department of Agricultural Products Quality and Safety Supervision Bureau of Agriculture Department, Ministry of Agriculture and Rural in China. Analysis and Summary of Unqualified Sampling by Edible Agricultural Products Market Supervision Authorities 8 in 2020; Department of Agricultural Products Quality and Safety Supervision Bureau of Agriculture Department, Ministry of Agriculture and Rural in China: Beijing, China, 2020.
- Tognaccini, L.; Ricci, M.; Gellini, C.; Feis, A.; Smulevich, G.; Becucci, M. Surface enhanced Raman spectroscopy for in-field detection of pesticides: A test on dimethoate residues in water and on olive leaves. Molecules 2019, 24, 292. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, R.; Geng, X.; Zhu, X.; Xiong, Y.; Chen, T.; Ai, S. Rapid detection of sulfonamide antibiotics residues in swine urine by surface-enhanced Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 15, 120570. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Chen, Y.; Xia, L.; Hu, Y.; Li, G. Flexible membrane composite based on sepiolite/chitosan/(silver nanoparticles) for enrichment and surface-enhanced Raman scattering determination of sulfamethoxazole in animal-derived food. Microchim. Acta 2022, 189, 199. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.E.; Goryacheva, I.Y.; Markin, A.V. SERS as a tool for determination of structurally related compounds: The case of sulfanilamide class antibiotics. Talanta 2024, 277, 126433. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.E.; Markin, A.V.; Weber, K.; Popp, J.; Cialla-May, D. Liquid-liquid extraction-assisted SERS-based determination of sulfamethoxazole in spiked human urine. Anal. Chim. Acta 2020, 1109, 61–68. [Google Scholar] [CrossRef] [PubMed]
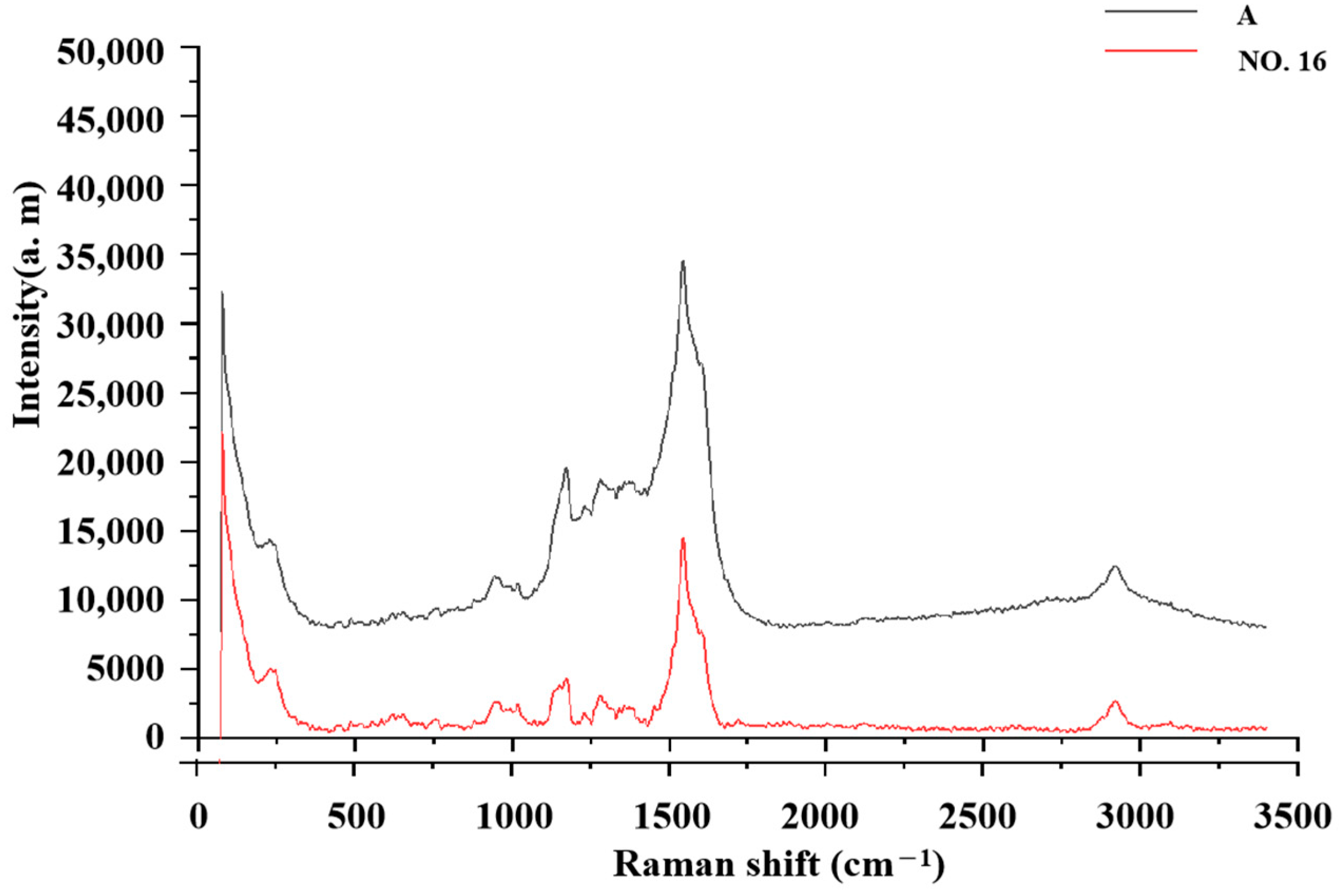
| Formula/Relative Rf | Raman Shift of the Blank Matrix (cm−1)/Relative Peak Intensity | SERS Shift (cm−1)/ Relative Intensity | Functional Group |
|---|---|---|---|
A 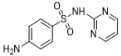 Rf = 1.20 | 3384~2999 (3 peaks) 1604~1104 (12 peaks) 1604/0.82 1510/0.15 1155/1.00 1104/0.34 | 3041~2802 (1 peaks) 1599~1018 (8 peaks) 1599/1.07 1544/1.54 1170/1.00 1091/0.31 | Common peaks: ν=CH, ν-CH2, ν-CH3 Characteristic peaks: νC=C from phenyl rings νC=C from phenyl rings νC-N νC-N |
B 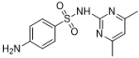 Rf = 1.57 | 3382~2858 (6 peaks) 1598~1096 (10 peaks) 1598/0.89 1513/0.12 1349/0.17 1149/1.00 1096/0.32 | 3035~2790 (1 peaks) 1578~1095 (7 peaks) 1578/2.80 1531/2.67 1356/1.51 1155/1.00 1095/0.30 | Common peaks: ν=CH, ν-CH2, ν-CH3 Characteristic peaks: νC=C from phenyl rings νC=C from phenyl rings βCH2, βCH3 νC-N νC-N |
C  Rf = 1.73 | 3381~2956 (6 peaks) 1604~1100 (9 peaks) 1604/0.57 1514/0.15 1312/0.13 1153/1.00 1100/0.35 | 3078~2790 (1 peaks) 1591~1082 (7 peaks) 1591/1.72 1550/2.07 1365/1.18 1155/1.00 1082/0.24 | Common peaks: ν=CH, ν-CH2, ν-CH3 Characteristic peaks: νC=C from phenyl rings νC=C from phenyl rings βCH2, βCH3 νC-N νC-N |
D  Rf = 1.63 | 3381~2946 (5 peaks) 1604~1100 (7 peaks) 1604/0.62 1515/0.18 1312/0.14 1154/1.00 1100/0.32 | 3064~2771 (1 peaks) 1592~1078 (6 peaks) 1578/2.45 1508/1.96 1318/1.10 1155/1.00 1096/0.24 | Common peaks: ν=CH, ν-CH2, ν-CH3 Characteristic peaks: νC=C from phenyl rings νC=C from phenyl rings βCH2, βCH3 νC-N νC-N |
E 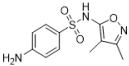 Rf = 1.00 | 3385~2938 (3 peaks) 1600~1097 (8 peaks) 1600/1.06 1502/0.41 1349/0.17 1158/1.00 1097/0.86 | 3055~2794 (1 peaks) 1576~1088 (6 peaks) 1573/2.31 1541/2.48 1338/1.52 1157/1.00 1088/0.39 | Common peaks: ν=CH, ν-CH2, ν-CH3 Characteristic peaks: νC=C from phenyl rings νC=C from phenyl rings βCH2, βCH3 νC-N νC-N |
F  Rf = 0.83 | 3297~3060 (3 peaks) 1601~1097 (8 peaks) 1601/1.09 1536/0.39 1138/1.00 1097/0.31 | 3041~2812 (1 peaks) 1545~1080 (10 peaks) 1568/2.15 1545/2.02 1151/1.00 1080/0.19 | Common peaks: ν=CH, ν-CH2, ν-CH3 Characteristic peaks: νC=C from phenyl rings νC=C from phenyl rings νC-N νC-N |
| Name | Functional Group | SERS | Raman | EF | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Raman Shift (cm−1) | ISERS | MSERS (µg) | ISERS/MSERS | Raman Shift (cm−1) | Iblank | Mblank (µg) | Iblank/Mblank | |||
| A | νC=C | 1606 | 11,761 | 4.0 × 10−3 | 2.9 × 10−6 | 1602 | 1540 | 10 | 154.0 | 1.9 × 104 |
| νC=C | 1523 | 9654 | 4.0 × 10−3 | 2.4 × 10−6 | 1513 | 465 | 10 | 46.5 | 5.2 × 104 | |
| βCH2, βCH3 | 1351 | 2506 | 4.0 × 10−3 | 6.3 × 10−5 | 1345 | 264 | 10 | 26.4 | 2.4 × 104 | |
| νC-N | 1158 | 4113 | 4.0 × 10−3 | 1.0 × 10−6 | 1154 | 213 | 10 | 21.3 | 4.8 × 104 | |
| νC-N | 1093 | 4200 | 4.0 × 10−3 | 1.1 × 10−6 | 1102 | 770 | 10 | 77.0 | 1.4 × 104 | |
| B | νC=C | 1579 | 10,035 | 4.0 × 10−3 | 2.5 × 10−6 | 1598 | 716 | 10 | 71.6 | 3.5 × 104 |
| νC=C | 1534 | 8516 | 4.0 × 10−3 | 2.1 × 10−6 | 1507 | 259 | 10 | 25.9 | 8.2 × 104 | |
| βCH2, βCH3 | 1376 | 2905 | 4.0 × 10−3 | 7.3 × 10−5 | 1349 | 524 | 10 | 52.4 | 1.4 × 104 | |
| νC-N | 1162 | 3111 | 4.0 × 10−3 | 7.8 × 10−5 | 1147 | 230 | 10 | 23.0 | 3.4 × 104 | |
| νC-N | 1088 | 905 | 4.0 × 10−3 | 2.3 × 10−5 | 1091 | 161 | 10 | 16.1 | 1.4 × 104 | |
| C | νC=C | 1596 | 10,134 | 4.0 × 10−3 | 2.5 × 10−6 | 1602 | 314 | 10 | 31.4 | 8.1 × 104 |
| νC=C | 1528 | 9365 | 4.0 × 10−3 | 2.3 × 10−6 | 1515 | 261 | 10 | 26.1 | 9.0 × 104 | |
| βCH2, βCH3 | 1372 | 3051 | 4.0 × 10−3 | 7.6 × 10−5 | 1357 | 519 | 10 | 51.9 | 1.5 × 104 | |
| νC-N | 1164 | 3731 | 4.0 × 10−3 | 9.3 × 10−5 | 1151 | 282 | 10 | 28.2 | 3.3 × 104 | |
| νC-N | 1089 | 4980 | 4.0 × 10−3 | 1.2 × 10−6 | 1098 | 288 | 10 | 28.8 | 4.3 × 104 | |
| D | νC=C | 1575 | 10,014 | 4.0 × 10−3 | 2.5 × 10−6 | 1600 | 610 | 10 | 61.0 | 4.1 × 104 |
| νC=C | 1532 | 8104 | 4.0 × 10−3 | 2.0 × 10−6 | 1506 | 220 | 10 | 22.0 | 9.2 × 104 | |
| βCH2, βCH3 | 1282 | 2412 | 4.0 × 10−3 | 6.0 × 10−5 | 1309 | 381 | 10 | 38.1 | 1.6 × 104 | |
| νC-N | 1168 | 3490 | 4.0 × 10−3 | 8.7 × 10−5 | 1158 | 217 | 10 | 21.7 | 4.0 × 104 | |
| νC-N | 1114 | 4510 | 4.0 × 10−3 | 1.1 × 10−6 | 1100 | 161 | 10 | 16.1 | 7.0 × 104 | |
| E | νC=C | 1590 | 18,901 | 4.0 × 10−3 | 4.7 × 10−6 | 1592 | 523 | 10 | 52.3 | 9.0 × 104 |
| νC=C | 1500 | 10,512 | 4.0 × 10−3 | 2.6 × 10−6 | 1498 | 83 | 10 | 8.3 | 3.2 × 105 | |
| βCH2, βCH3 | 1326 | 1320 | 4.0 × 10−3 | 3.3 × 10−5 | 1318 | 289 | 10 | 28.9 | 1.1 × 104 | |
| νC-N | 1158 | 8645 | 4.0 × 10−3 | 2.2 × 10−6 | 1164 | 128 | 10 | 12.8 | 1.7 × 105 | |
| νC-N | 1097 | 1693 | 4.0 × 10−3 | 4.2 × 10−5 | 1093 | 197 | 10 | 19.7 | 2.1 × 104 | |
| F | νC=C | 1582 | 14,423 | 4.0 × 10−3 | 3.6 × 10−6 | 1588 | 367 | 10 | 36.7 | 9.8 × 104 |
| νC=C | 1538 | 17,891 | 4.0 × 10−3 | 4.5 × 10−6 | 1540 | 188 | 10 | 18.8 | 2.4 × 105 | |
| βCH2, βCH3 | 1363 | 3597 | 4.0 × 10−3 | 9.0 × 10−5 | 1330 | 225 | 10 | 22.5 | 4.0 × 104 | |
| νC-N | 1151 | 7245 | 4.0 × 10−3 | 1.8 × 10−6 | 1149 | 105 | 10 | 10.5 | 1.7 × 105 | |
| νC-N | 1073 | 1728 | 4.0 × 10−3 | 4.3 × 10−5 | 1097 | 217 | 10 | 21.7 | 2.0 × 104 | |
| Compound | MRL’ (μg/kg) | LOD’ (μg/kg) | MRL (ng/mL) | LOD (ng/mL) |
|---|---|---|---|---|
| Sulfamethasone (A) | 100.0 | 3.1 | 400.0 | 12.5 |
| Sulfamethazine (B) | 100.0 | 1.6 | 400.0 | 6.4 |
| Sulfadoxine (C) | 100.0 | 1.6 | 400.0 | 6.3 |
| Sulfamethoxydiazine (D) | 100.0 | 1.8 | 400.0 | 7.1 |
| Sulfamethoxazole (E) | 100.0 | 4.7 | 400.0 | 18.8 |
| Sulfathiazole (F) | 100.0 | 1.6 | 400.0 | 6.2 |
| Compound | Characteristic Peak | Peak | Peak Intensity | RSD% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 3 h | 6 h | 9 h | 12 h | 24 h | 48 h | ||||
| A | νC=C | 1589 | 12,378 | 12,409 | 12,433 | 12,298 | 12,192 | 12,385 | 12,180 | 0.8 |
| νC=C | 1541 | 20,387 | 20,386 | 20,451 | 20,299 | 20,261 | 20,189 | 20,377 | 0.4 | |
| νC-N | 1170 | 13,238 | 13,301 | 13,221 | 13,298 | 13,212 | 13,245 | 13,210 | 0.3 | |
| νC-N | 1094 | 4101 | 4067 | 3950 | 4043 | 4131 | 4043 | 4013 | 1.5 | |
| B | νC=C | 1579 | 30,482 | 30,501 | 30,512 | 30,497 | 30,498 | 30,677 | 30,479 | 0.2 |
| νC=C | 1527 | 29,067 | 29,087 | 29,101 | 29,087 | 29,076 | 29,054 | 29,081 | 0.1 | |
| βCH2, βCH3 | 1353 | 16,438 | 16,442 | 16,428 | 16,451 | 16,387 | 16,459 | 16,432 | 0.1 | |
| νC-N | 1151 | 10,886 | 10,874 | 10,890 | 10,881 | 10,876 | 10,873 | 10,898 | 0.1 | |
| νC-N | 1096 | 3266 | 3247 | 3259 | 3275 | 3269 | 3184 | 3190 | 1.2 | |
| C | νC=C | 1593 | 26,710 | 26,687 | 26,736 | 26,692 | 26,714 | 26,721 | 26,698 | 0.1 |
| νC=C | 1551 | 32,145 | 32,157 | 32,210 | 32,152 | 32,163 | 32,171 | 32,156 | 0.1 | |
| βCH2, βCH3 | 1367 | 18,324 | 18,318 | 18,319 | 18,325 | 18,301 | 18,327 | 18,330 | 0.1 | |
| νC-N | 1160 | 15,529 | 15,527 | 15,531 | 15,537 | 15,547 | 15,534 | 15,512 | 0.1 | |
| νC-N | 1082 | 3727 | 3736 | 3743 | 3729 | 3736 | 3721 | 3719 | 0.2 | |
| D | νC=C | 1573 | 34,198 | 34,210 | 34,201 | 34,199 | 34,209 | 34,211 | 34,312 | 0.1 |
| νC=C | 1510 | 27,356 | 27,362 | 27,340 | 27,367 | 27,382 | 27,374 | 27,396 | 0.1 | |
| βCH2, βCH3 | 1316 | 15,354 | 15,462 | 15,341 | 15,367 | 15,344 | 15,361 | 15,349 | 0.3 | |
| νC-N | 1154 | 13,958 | 13,966 | 13,973 | 13,881 | 13,941 | 13,970 | 14,163 | 0.6 | |
| νC-N | 1097 | 3350 | 3353 | 3367 | 3341 | 3226 | 3359 | 3461 | 2.0 | |
| E | νC=C | 1573 | 18,711 | 18,701 | 18,698 | 18,687 | 18,723 | 18,691 | 18,707 | 0.1 |
| νC=C | 1542 | 20,088 | 20,095 | 20,067 | 20,087 | 20,075 | 20,074 | 20,064 | 0.1 | |
| βCH2, βCH3 | 1339 | 12,312 | 12,269 | 12,328 | 12,300 | 12,268 | 12,276 | 12,293 | 0.2 | |
| νC-N | 1161 | 8120 | 7969 | 8128 | 7900 | 8168 | 8176 | 7993 | 1.4 | |
| νC-N | 1087 | 3160 | 3169 | 3128 | 3170 | 3168 | 3176 | 3189 | 0.6 | |
| F | νC=C | 1560 | 41,031 | 40,976 | 40,986 | 40,989 | 41,026 | 41,028 | 41,020 | 0.1 |
| νC=C | 1544 | 38,549 | 38,551 | 38,539 | 38,543 | 38,611 | 38,563 | 38,557 | 0.1 | |
| νC-N | 1149 | 19,084 | 19,089 | 19,094 | 19,076 | 19,079 | 19,187 | 19,093 | 0.2 | |
| νC-N | 1076 | 3626 | 3639 | 3617 | 3620 | 3649 | 3623 | 3645 | 0.4 | |
| Compound | Characteristic Peak | Peak | Intensity 1 | Intensity 2 | Intensity 3 | RSD% |
|---|---|---|---|---|---|---|
| A | νC-N | 1094 | 7561 | 7503 | 7693 | 1.3 |
| νC-N | 1096 | 4709 | 4740 | 4801 | 1.0 | |
| νC-N | 1094 | 4104 | 4177 | 4113 | 1.0 | |
| B | νC-N | 1098 | 6521 | 6504 | 6374 | 1.2 |
| νC-N | 1100 | 4636 | 4667 | 4604 | 0.7 | |
| νC-N | 1096 | 3266 | 3191 | 3154 | 1.8 | |
| C | νC-N | 1178 | 4073 | 3985 | 3957 | 1.5 |
| νC-N | 1180 | 3849 | 3770 | 3770 | 1.2 | |
| νC-N | 1182 | 3727 | 3702 | 3562 | 2.4 | |
| D | νC-N | 1096 | 10,158 | 10,368 | 10,220 | 1.1 |
| νC-N | 1098 | 4814 | 4741 | 4744 | 0.9 | |
| νC-N | 1097 | 3350 | 3242 | 3243 | 1.9 | |
| E | νC-N | 1084 | 8005 | 8165 | 7840 | 2.0 |
| νC-N | 1092 | 5973 | 5758 | 5882 | 1.8 | |
| νC-N | 1087 | 3159 | 3206 | 3149 | 1.0 | |
| F | νC-N | 1081 | 8073 | 7830 | 7951 | 1.5 |
| νC-N | 1079 | 5036 | 5237 | 5183 | 2.0 | |
| νC-N | 1076 | 3626 | 3611 | 3687 | 1.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Zhang, M.; Li, L.; Liu, J.; Xu, F.; Wang, Y.; Song, B.; Xu, T.; Hong, Y.; Zhang, H. Study on the Rapid Limit Test for Six Sulfonamide Residues in Food Based on the TLC-SERS Method. Molecules 2024, 29, 3977. https://doi.org/10.3390/molecules29163977
Ma Y, Zhang M, Li L, Liu J, Xu F, Wang Y, Song B, Xu T, Hong Y, Zhang H. Study on the Rapid Limit Test for Six Sulfonamide Residues in Food Based on the TLC-SERS Method. Molecules. 2024; 29(16):3977. https://doi.org/10.3390/molecules29163977
Chicago/Turabian StyleMa, Yukun, Min Zhang, Li Li, Jicheng Liu, Feng Xu, Yuanrui Wang, Bo Song, Tao Xu, Yue Hong, and Honglian Zhang. 2024. "Study on the Rapid Limit Test for Six Sulfonamide Residues in Food Based on the TLC-SERS Method" Molecules 29, no. 16: 3977. https://doi.org/10.3390/molecules29163977
APA StyleMa, Y., Zhang, M., Li, L., Liu, J., Xu, F., Wang, Y., Song, B., Xu, T., Hong, Y., & Zhang, H. (2024). Study on the Rapid Limit Test for Six Sulfonamide Residues in Food Based on the TLC-SERS Method. Molecules, 29(16), 3977. https://doi.org/10.3390/molecules29163977







