Effects of Isoxazolyl Steroids on Key Genes of Sonic Hedgehog Cascade Expression in Tumor Cells
Abstract
:1. Introduction
2. Results
2.1. Docking Results
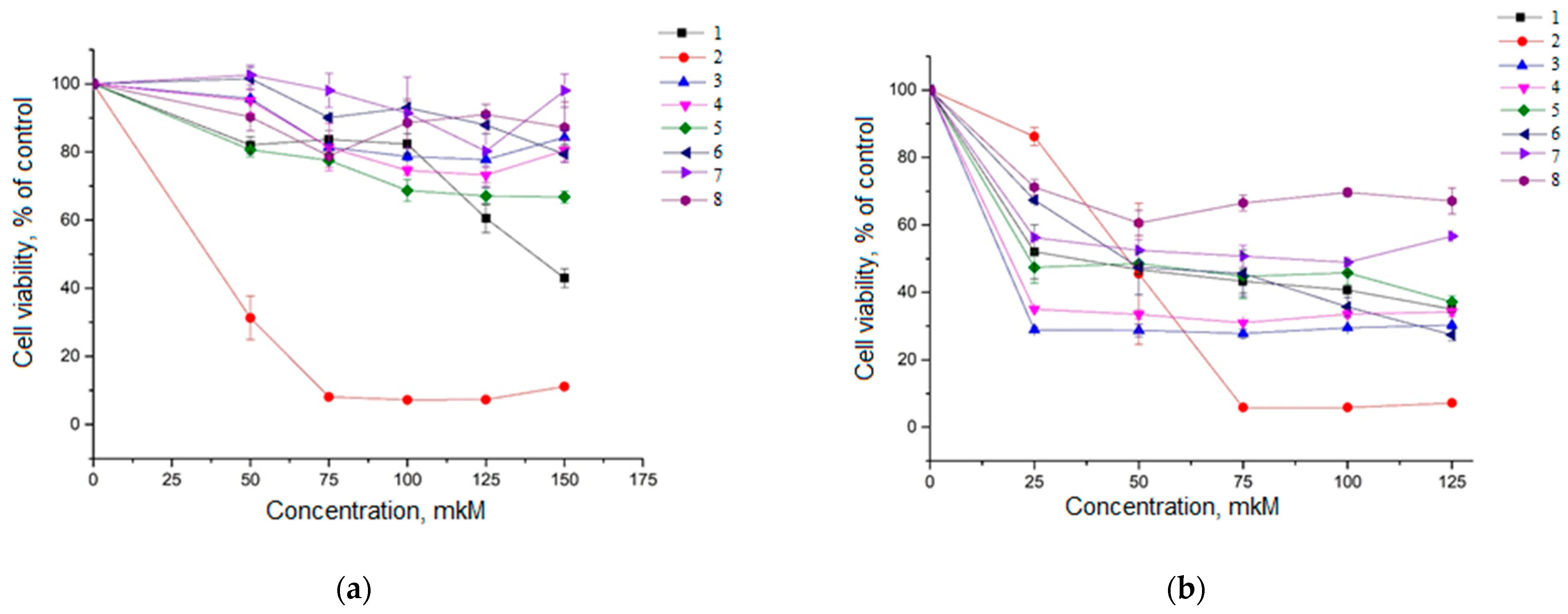
2.2. Cytotoxicity Analysis of Compounds 1–8 in HeLa Cells and A549 Cells
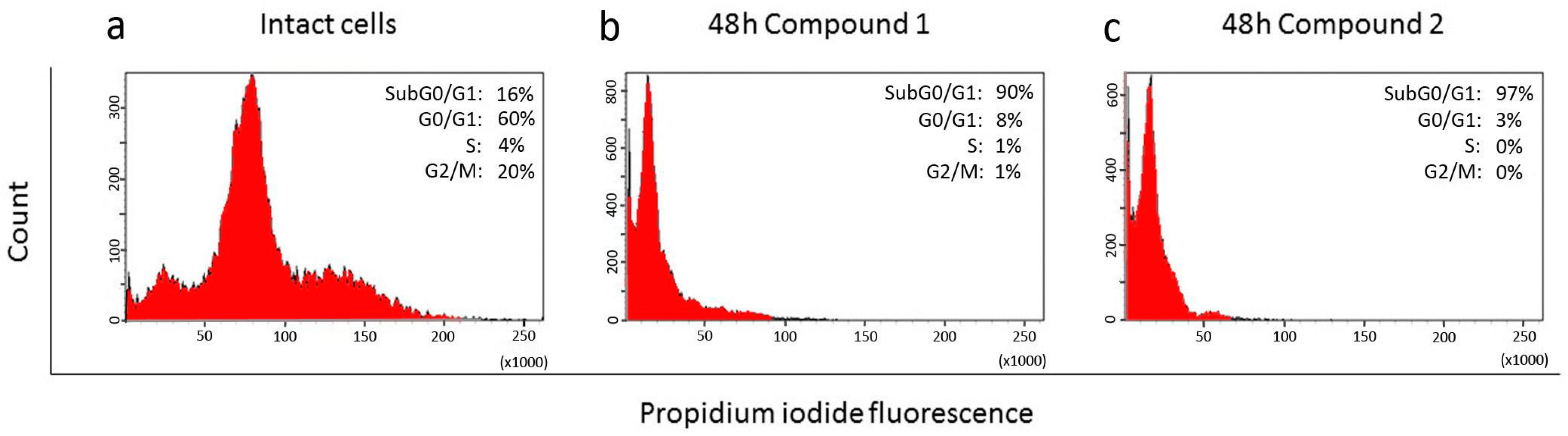
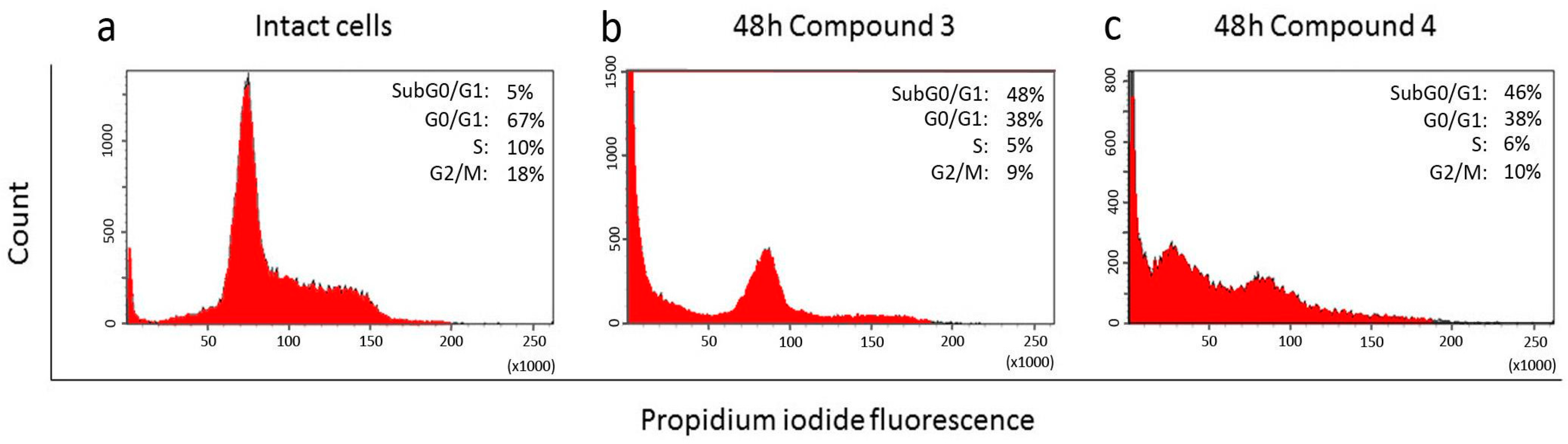
2.3. Effect of Compounds 1, 2, 3, and 4 on the Cell Cycle
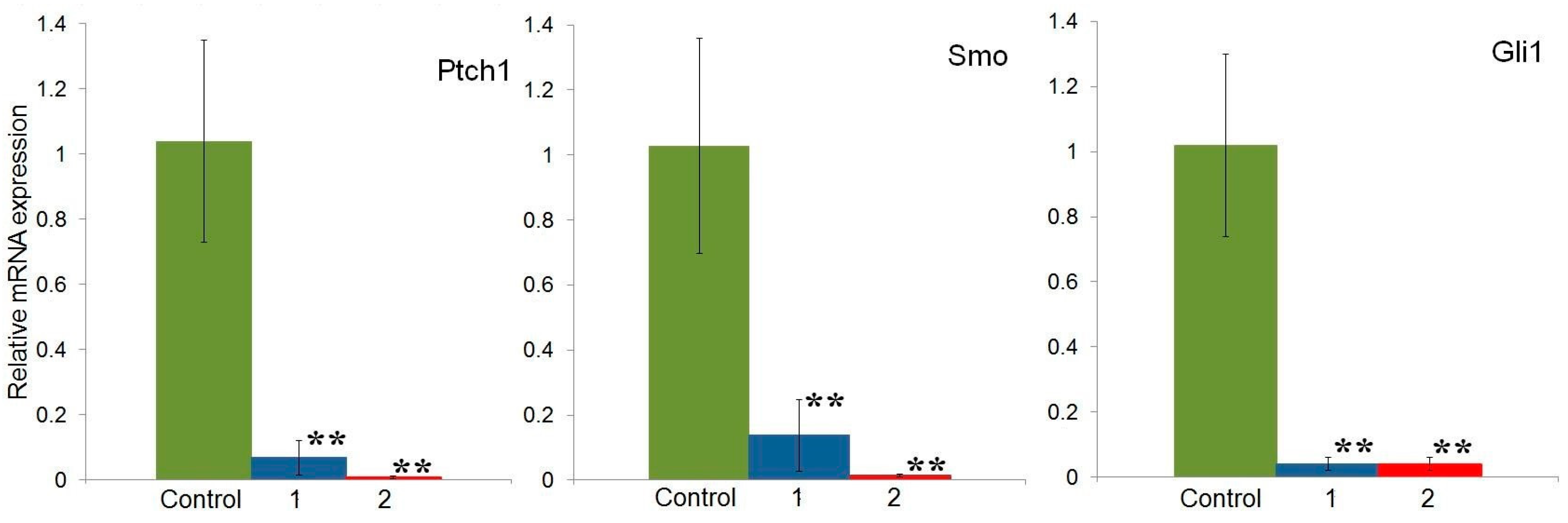
2.4. Effect of Compounds 1 and 2 on the Expression of Hh Signaling Pathway Genes in HeLa Cells
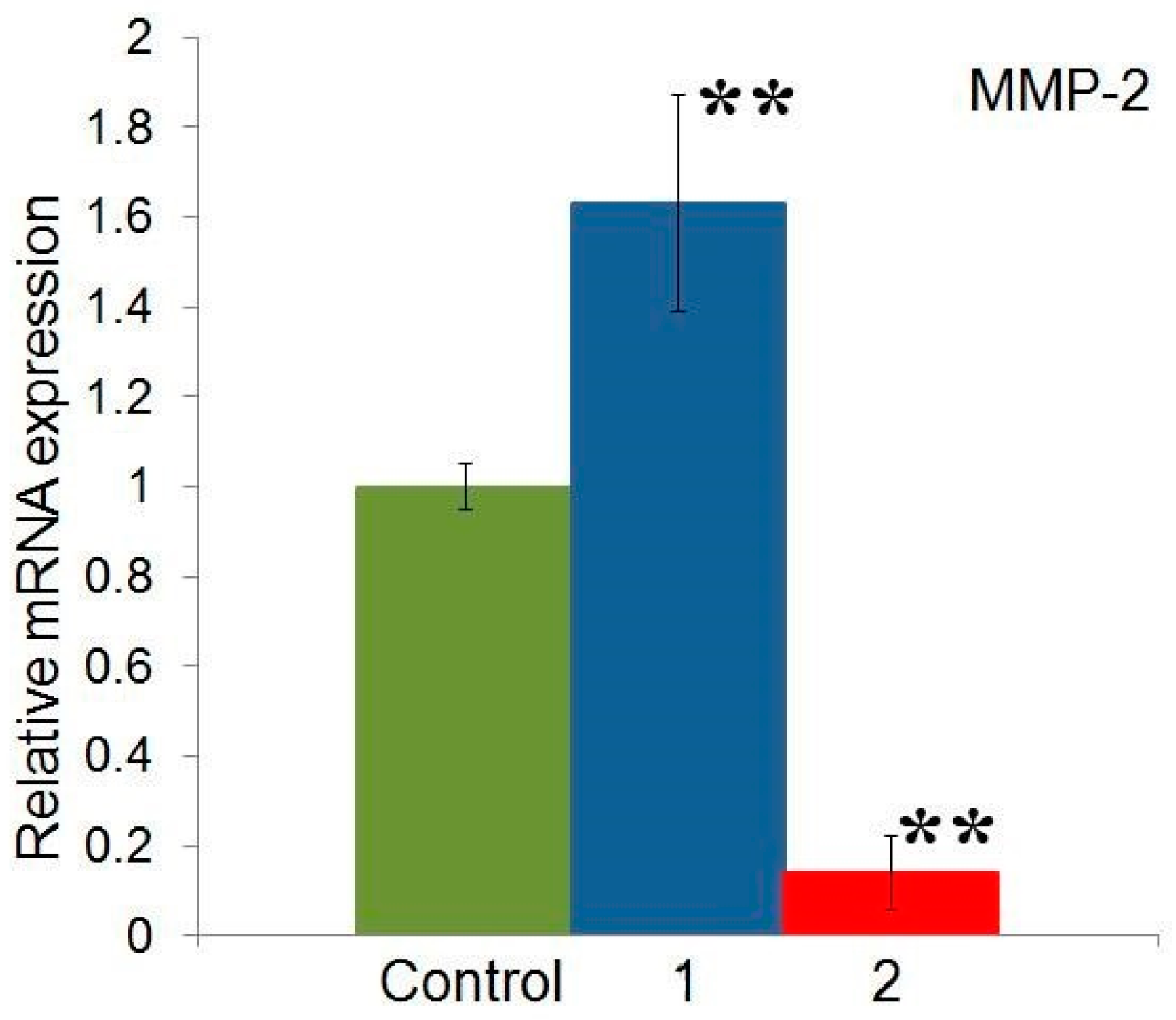
2.5. Effect of Compounds 1 and 2 on the Expression of MMP-2 Gene in HeLa Cells
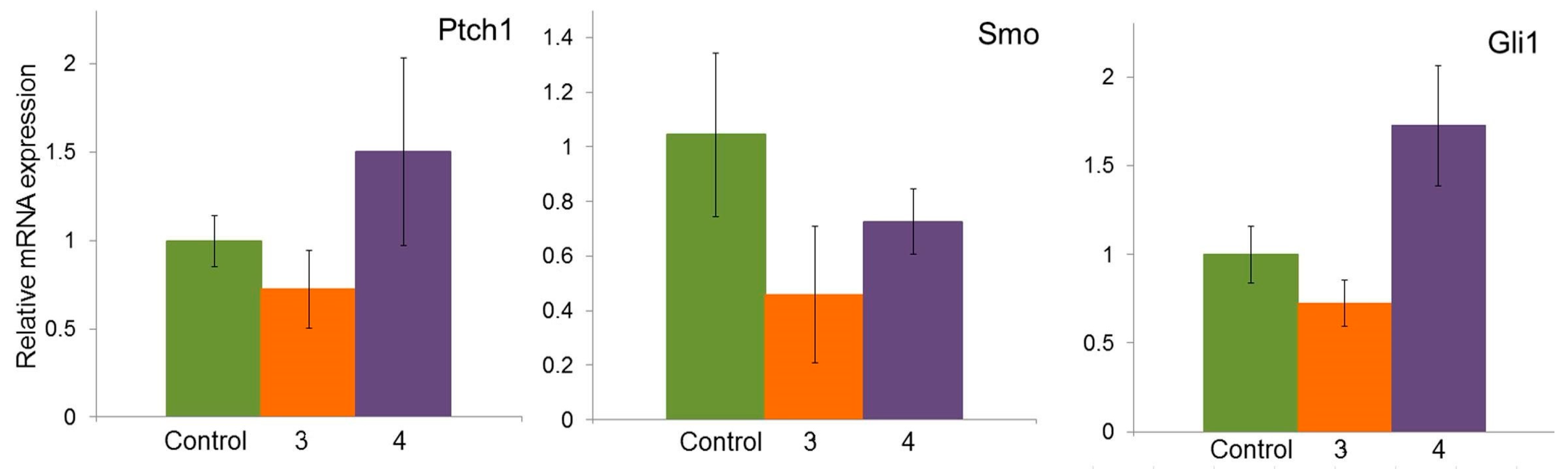
2.6. Effect of Compounds 3 and 4 on the Expression of Hh Signaling Pathway Genes in A549 Cells
2.7. Effect of Compounds 3 and 4 on the Gene Expression of MMP-2 and MMP-9 in A549 Cells
2.8. Effect of Compounds 1 and 2 on Proliferation in the MCF-7 Cell Line
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Molecular Docking
4.3. Cells and Culture
4.4. Cytotoxicity Evaluation (MTT Test)
4.5. Cell Cycle Analysis
4.6. Real-Time PCR
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AR | androgen receptor |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DMSO | dimethylsulfoxide |
| ECM | extracellular matrix |
| ER | estrogen receptor |
| FBS | fetal bovine serum |
| Hh | Hedgehog pathway |
| MMPs | metalloproteinases |
| PBS | phosphate-buffered saline |
| PI | propidium iodide |
| RMSD | root-mean-square deviation |
| Smo | The protein that carries the Hh signal across the membrane is the oncoprotein and G-protein coupled receptor (GPCR) Smoothened (Smo) |
References
- Wu, F.; Zhang, Y.; Sun, B.; McMahon, A.P.; Wang, Y. Hedgehog signaling: From basic biology to cancer therapy. Cell Chem. Biol. 2017, 24, 252–280. [Google Scholar] [CrossRef]
- Skoda, A.M.; Simovic, D.; Karin, V.; Kardum, V.; Vranic, S.; Serman, L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018, 18, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hsia, E.Y.; Brigui, A.; Plessis, A.; Beachy, P.A.; Zheng, X. The role of ciliary trafficking in Hedgehog receptor signaling. Sci. Signal. 2015, 8, ra55. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, H.; Liu, A. The loss of Hh responsiveness by a non-ciliary Gli2 variant. Development 2015, 142, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, Z.; Zhan, X.; Cao, M.; Satoh, T.; Akira, S.; Shpargel, K.; Magnuson, T.; Li, Q.; Wang, R.; et al. An epigenetic switch induced by Shh signalling regulates gene activation during development and medulloblastoma growth. Nat. Commun. 2014, 5, 5425. [Google Scholar] [CrossRef] [PubMed]
- Junker, J.P.; Peterson, K.A.; Nishi, Y.; Mao, J.; McMahon, A.P.; van Oudenaarden, A. A predictive model of bifunctional transcription factor signaling during embryonic tissue patterning. Dev. Cell 2014, 31, 448–460. [Google Scholar] [CrossRef]
- Peterson, K.A.; Nishi, Y.; Ma, W.; Vedenko, A.; Shokri, L.; Zhang, X.; McFarlane, M.; Baizabal, J.M.; Junker, J.P.; van Oudenaarden, A.; et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012, 26, 2802–2816. [Google Scholar] [CrossRef]
- Hu, Z.; Xie, F.; Hu, A.; Xu, M.; Liu, Y.; Zhang, J.; Xiao, J.; Song, Y.; Zhong, J.; Chen, B. Silencing glioma-associated oncogene homolog 1 suppresses the migration and invasion of hepatocellular carcinoma in vitro. Oncol. Lett. 2020, 20, 228. [Google Scholar] [CrossRef]
- Garcia, N.; Ulin, M.; Al-Hendy, A.; Yang, Q. The role of hedgehog pathway in female cancers. J. Cancer Sci. Clin. Ther. 2020, 4, 487–498. [Google Scholar] [CrossRef]
- Cabral-Pacheco, G.A.; Garza-Veloz, I.; Castruita-De la Rosa, C.; Ramirez-Acuña, J.M.; Perez-Romero, B.A.; Guerrero-Rodriguez, J.F.; Martinez-Avila, N.; Martinez-Fierro, M.L. The roles of matrix metalloproteinases and their inhibitors in human diseases. Int. J. Mol. Sci. 2020, 21, 9739. [Google Scholar] [CrossRef]
- Bosch, R.; Philips, N.; Suárez-Pérez, J.A.; Juarranz, A.; Devmurari, A.; Chalensouk-Khaosaat, J.; González, S. Mechanisms of Photoaging and Cutaneous Photocarcinogenesis, and Photoprotective Strategies with Phytochemicals. Antioxidants 2015, 4, 248–268. [Google Scholar] [CrossRef] [PubMed]
- Azevedo Martins, J.M.; Rabelo-Santos, S.H.; do Amaral Westin, M.C.; Zeferino, L.C. Tumoral and stromal expression of MMP-2, MMP-9, MMP-14, TIMP-1, TIMP-2, and VEGF-A in cervical cancer patient survival: A competing risk analysis. BMC Cancer 2020, 20, 660. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Maguire, T.M.; Hill, A.; McDermott, E.; O’Higgins, N. Metalloproteinases: Role in breast carcinogenesis, invasion and metastasis. Breast Cancer Res. 2000, 2, 252–257. [Google Scholar] [CrossRef]
- Solovyeva, N.I.; Timoshenko, O.S.; Gureeva, T.A.; Kugaevskaya, E.V. Matrix metalloproteinases and their endogenous regulators in squamous cervical carcinoma (review of the own data). Biomed. Khimiya 2015, 61, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Li, X.; Gao, Y.; Li, Y.; Yang, P.; Wang, K. Identification and validation of the role of matrix metalloproteinase-1 in cervical cancer. Int. J. Oncol. 2018, 52, 1198–1208. [Google Scholar] [CrossRef]
- Zhu, D.; Ye, M.; Zhang, W. E6/E7 oncoproteins of high risk HPV-16 upregulate MT1-MMP, MMP-2 and MMP-9 and promote the migration of cervical cancer cells. Int. J. Clin. Exp. Pathol. 2015, 8, 4981–4989. [Google Scholar]
- Rudovich, A.S.; Peřina, M.; Krech, A.V.; Novozhilova, M.Y.; Tumilovich, A.M.; Shkel, T.V.; Grabovec, I.P.; Kvasnica, M.; Mada, L.; Zavialova, M.G.; et al. Synthesis and biological evaluation of new isoxazolyl steroids as anti-prostate cancer agents. Int. J. Mol. Sci. 2022, 23, 13534. [Google Scholar] [CrossRef]
- Giroux-Leprieur, E.; Costantini, A.; Ding, V.W.; He, B. Hedgehog signaling in lung cancer: From oncogenesis to cancer treatment resistance. Int. J. Mol. Sci. 2018, 19, 2835. [Google Scholar] [CrossRef]
- Samarzija, I.; Beard, P. Hedgehog pathway regulators influence cervical cancer cell proliferation, survival and migration. Biochem. Biophys. Res. Commun. 2012, 425, 64–69. [Google Scholar] [CrossRef]
- Roomi, M.W.; Monterrey, J.C.; Kalinovsky, T.; Rath, M.; Niedzwiecki, A. In vitro modulation of MMP-2 and MMP-9 in human cervical and ovarian cancer cell lines by cytokines, inducers and inhibitors. Oncol. Rep. 2010, 23, 605–614. [Google Scholar] [CrossRef]
- Schröpfer, A.; Kammerer, U.; Kapp, M.; Dietl, J.; Feix, S.; Anacker, J. Expression pattern of matrix metalloproteinases in human gynecological cancer cell lines. BMC Cancer 2010, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Madeddu, L.; Legros, N.; Devleeschouwer, N.; Bosman, C.; Piccart, M.J.; LeClercq, G. Estrogen receptor status and estradiol sensitivity of MCF-7 cells in exponential growth phase. Eur. J. Cancer Clin. Oncol. 1988, 24, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Sheng, T.; Li, C.; Zhang, X.; Chi, S.; He, N.; Chen, K.; McCormick, F.; Gatalica, Z.; Xie, J. Activation of the hedgehog pathway in advanced prostate cancer. Mol. Cancer 2004, 3, 29. [Google Scholar] [CrossRef]
- Bailey, J.M.; Mohr, A.M.; Hollingsworth, M.A. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene 2009, 28, 3513–3525. [Google Scholar] [CrossRef]
- Ma, X.; Sheng, T.; Zhang, Y.; Zhang, X.; He, J.; Huang, S.; Chen, K.; Sultz, J.; Adegboyega, P.A.; Zhang, H.; et al. Hedgehog signaling is activated in subsets of esophageal cancers. Int. J. Cancer 2006, 118, 139–148. [Google Scholar] [CrossRef]
- Chaudary, N.; Pintilie, M.; Hedley, D.; Fyles, A.W.; Milosevic, M.; Clarke, B.; Hill, R.P.; Mackay, H. Hedgehog pathway signaling in cervical carcinoma and outcome after chemoradiation. Cancer 2012, 118, 3105–3115. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Jung, H.S.; Choi, Y.L.; Shin, Y.K.; Kim, H.J.; Kim, K.H.; Kim, W.J.; Lee, Y.J.; Kim, S.H. Enhanced expression of hedgehog signaling molecules in squamous cell carcinoma of uterine cervix and its precursor lesions. Mod. Pathol. 2006, 19, 1139–1147. [Google Scholar] [CrossRef]
- Yue, D.; Li, H.; Che, J.; Zhang, Y.; Tseng, H.H.; Jin, J.Q.; Luh, T.M.; Giroux-Leprieur, E.; Mo, M.; Zheng, Q.; et al. Hedgehog/Gli promotes epithelial-mesenchymal transition in lung squamous cell carcinomas. J. Exp. Clin. Cancer Res. 2014, 33, 34. [Google Scholar] [CrossRef]
- Singh, R.; Dhanyamraju, P.K.; Lauth, M. DYRK1B blocks canonical and promotes non-canonical Hedgehog signaling through activation of the mTOR/AKT pathway. Oncotarget 2017, 8, 833–845. [Google Scholar] [CrossRef]
- Verma, R.P.; Hansch, C. Matrix metalloproteinases (MMPs): Chemical-biological functions and (Q)SARs. Bioorg. Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yu, K.; Lian, Z.; Deng, S. Molecular regulation of androgen receptors in major female reproductive system cancers. Int. J. Mol. Sci. 2022, 23, 7556. [Google Scholar] [CrossRef]
- Noël, J.C.; Bucella, D.; Fayt, I.; Simonart, T.; Buxant, F.; Anaf, V.; Simon, P. Androgen receptor expression in cervical intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix. Int. J. Gynecol. Pathol. 2008, 27, 437–441. [Google Scholar] [CrossRef]
- Lykkesfeldt, A.E.; Sørensen, E.K. Effect of estrogen and antiestrogens on cell proliferation and synthesis of secreted proteins in the human breast cancer cell line MCF-7 and a tamoxifen resistant variant subline, AL-1. Acta Oncol. 1992, 31, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.R.; Moon, J.Y.; Ediriweera, M.K.; Song, Y.W.; Cho, M.; Kasiviswanathan, D.; Cho, S.K. Dietary flavonoid myricetin inhibits invasion and migration of radioresistant lung cancer cells (A549-IR) by suppressing MMP-2 and MMP-9 expressions through inhibition of the FAK-ERK signaling pathway. Food Sci. Nutr. 2020, 8, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; He, D.; Zhao, S.; Wang, H. IL-17A/IL-17RA promotes invasion and activates MMP-2 and MMP-9 expression via p38 MAPK signaling pathway in non-small cell lung cancer. Mol. Cell. Biochem. 2019, 455, 195–206. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(−delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]



| Compound | Hydrophobic Interactions | H-Bonds | Average Affinity for Cluster (kcal/mol) | |
|---|---|---|---|---|
| Residue | Length, Å | |||
| Cyclopamine | F221, L303, L384, V386, Y394, F484, P513 | E518 | 2.7 | - |
| 1 | I215, L221, F484, P513 | Y207 | 3.7 | |
| E208 | 3.0 | |||
| E208 | 3.4 | |||
| K395 | 2.9 | −10.9 | ||
| D473 | 2.7 | |||
| 2 | Y207, E208, L303, Y394, W480, F484, P513 | Y207 | 3.0 | |
| K395 | 2.6 | −10.7 | ||
| 3 | Y207, E208, L221, L303, Y394, W480, F484, P513 | K395 | 2.8 | −12.1 |
| 4 | Y207, E208, I215, L303, Y394, W480, F484 | Y207 | 3.7 | |
| K395 | 2.7 | |||
| R480 | 3.6 | |||
| R480 | 3.1 | −12.1 | ||
| D473 | 3.1 | |||
| 5 | Y207, Y394, W480, F484 | Y207 | 4.0 | −10.1 |
| K395 | 2.9 | |||
| D473 | 2.6 | |||
| D473 | 4.1 | |||
| 6 | Y204, E208, I215, L221, L303, Y394, F484, P513 | Y207 | 3.7 | −11.1 |
| K395 | 3.6 | |||
| 7 | L221, L303, F484, P513 | K384 | 2.5 | −10.6 |
| R480 | 3.9 | |||
| R480 | 3.8 | |||
| D473 | 2.7 | |||
| 8 | F484, P513 | Y207 | 3.8 | −10.5 |
| K395 | 3.2 | |||
| E481 | 3.5 | |||
| No | Name | Structure |
|---|---|---|
| 1 | 17β-((3-(2-hydroxypropan-2-yl)isoxazol-5-yl)methyl)androst-5-en-3β-ol |  |
| 2 | 17β-((3-butylisoxazol-5-yl)methyl)androst-5-en-3β-ol |  |
| 3 | 3β-hydroxy-17β-((3-(pyridin-2-yl)isoxazol-5-yl)methyl)androst-5-ene | 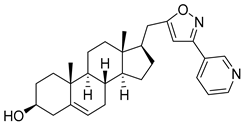 |
| 4 | (17β)-17-((3-phenylisoxazol-5-yl)methyl)androst-5-en-3β-ol |  |
| 5 | (E)-1-((17R)-3β-hydroxyandrost-5-en-17-yl)-4-(methoxy(methyl)amino)but-3-en-2-one | 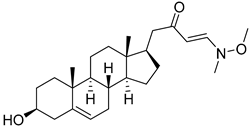 |
| 6 | 3-cyclopropyl-5-(((17R)-3β-hydroxyandrost-5-en-17-yl)methyl)-4,5-dihydroisoxazol-5-ol | 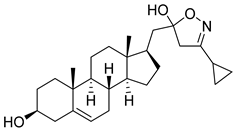 |
| 7 | (17R)-17-((3-(chloromethyl)isoxazol-5-yl)methyl)androst-5-en-3β-ol | 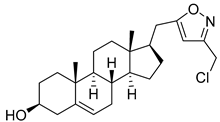 |
| 8 | 17β-((3-(hydroxymethyl)isoxazol-5-yl)methyl)androst-5-en-3β-ol |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleksandrova, A.; Mekhtiev, A.; Timoshenko, O.; Kugaevskaya, E.; Gureeva, T.; Gisina, A.; Zavialova, M.; Scherbakov, K.; Rudovich, A.; Zhabinskii, V.; et al. Effects of Isoxazolyl Steroids on Key Genes of Sonic Hedgehog Cascade Expression in Tumor Cells. Molecules 2024, 29, 4026. https://doi.org/10.3390/molecules29174026
Aleksandrova A, Mekhtiev A, Timoshenko O, Kugaevskaya E, Gureeva T, Gisina A, Zavialova M, Scherbakov K, Rudovich A, Zhabinskii V, et al. Effects of Isoxazolyl Steroids on Key Genes of Sonic Hedgehog Cascade Expression in Tumor Cells. Molecules. 2024; 29(17):4026. https://doi.org/10.3390/molecules29174026
Chicago/Turabian StyleAleksandrova, Anna, Arif Mekhtiev, Olga Timoshenko, Elena Kugaevskaya, Tatiana Gureeva, Alisa Gisina, Maria Zavialova, Kirill Scherbakov, Anton Rudovich, Vladimir Zhabinskii, and et al. 2024. "Effects of Isoxazolyl Steroids on Key Genes of Sonic Hedgehog Cascade Expression in Tumor Cells" Molecules 29, no. 17: 4026. https://doi.org/10.3390/molecules29174026
APA StyleAleksandrova, A., Mekhtiev, A., Timoshenko, O., Kugaevskaya, E., Gureeva, T., Gisina, A., Zavialova, M., Scherbakov, K., Rudovich, A., Zhabinskii, V., & Khripach, V. (2024). Effects of Isoxazolyl Steroids on Key Genes of Sonic Hedgehog Cascade Expression in Tumor Cells. Molecules, 29(17), 4026. https://doi.org/10.3390/molecules29174026









