Investigation on the Synergy between Membrane Permeabilizing Amphiphilic α-Hydrazido Acids and Commonly Used Antibiotics against Drug-Resistant Bacteria
Abstract
:1. Introduction
2. Results and Discussion
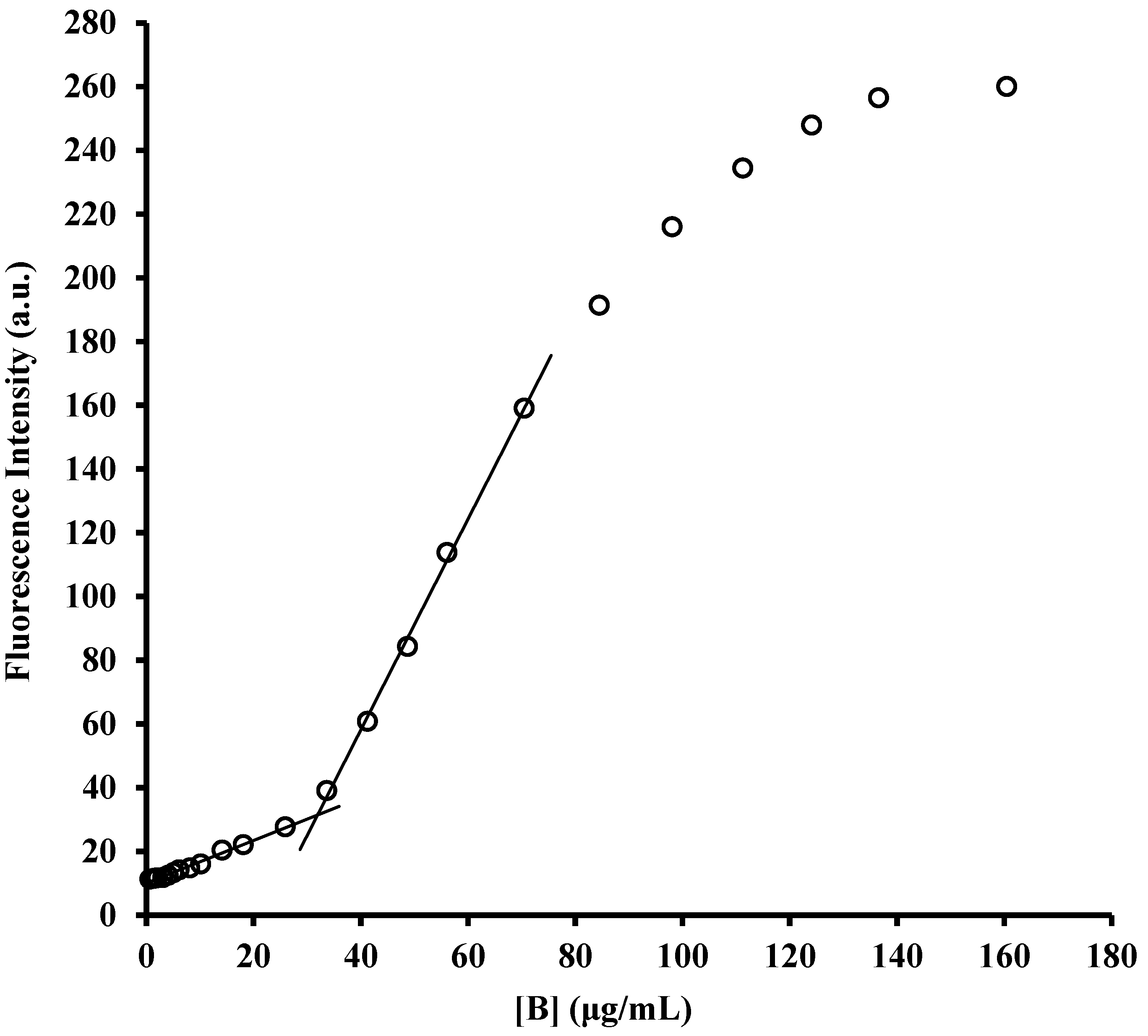
2.1. Determination of Critical Micellar Concentrations (CMC)
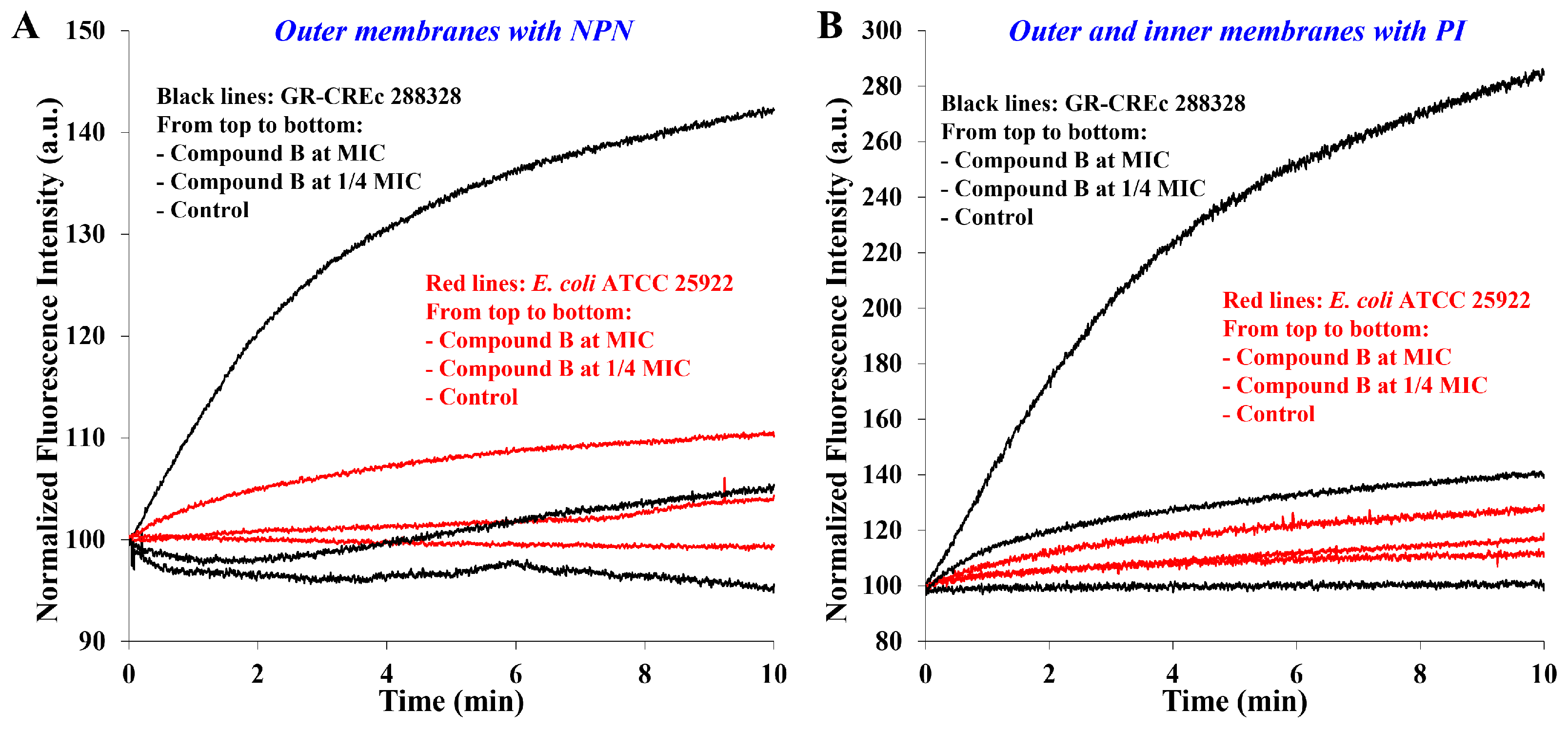
2.2. Membrane Permeabilization at Subtoxic Concentrations of α-Hydrazido Acids
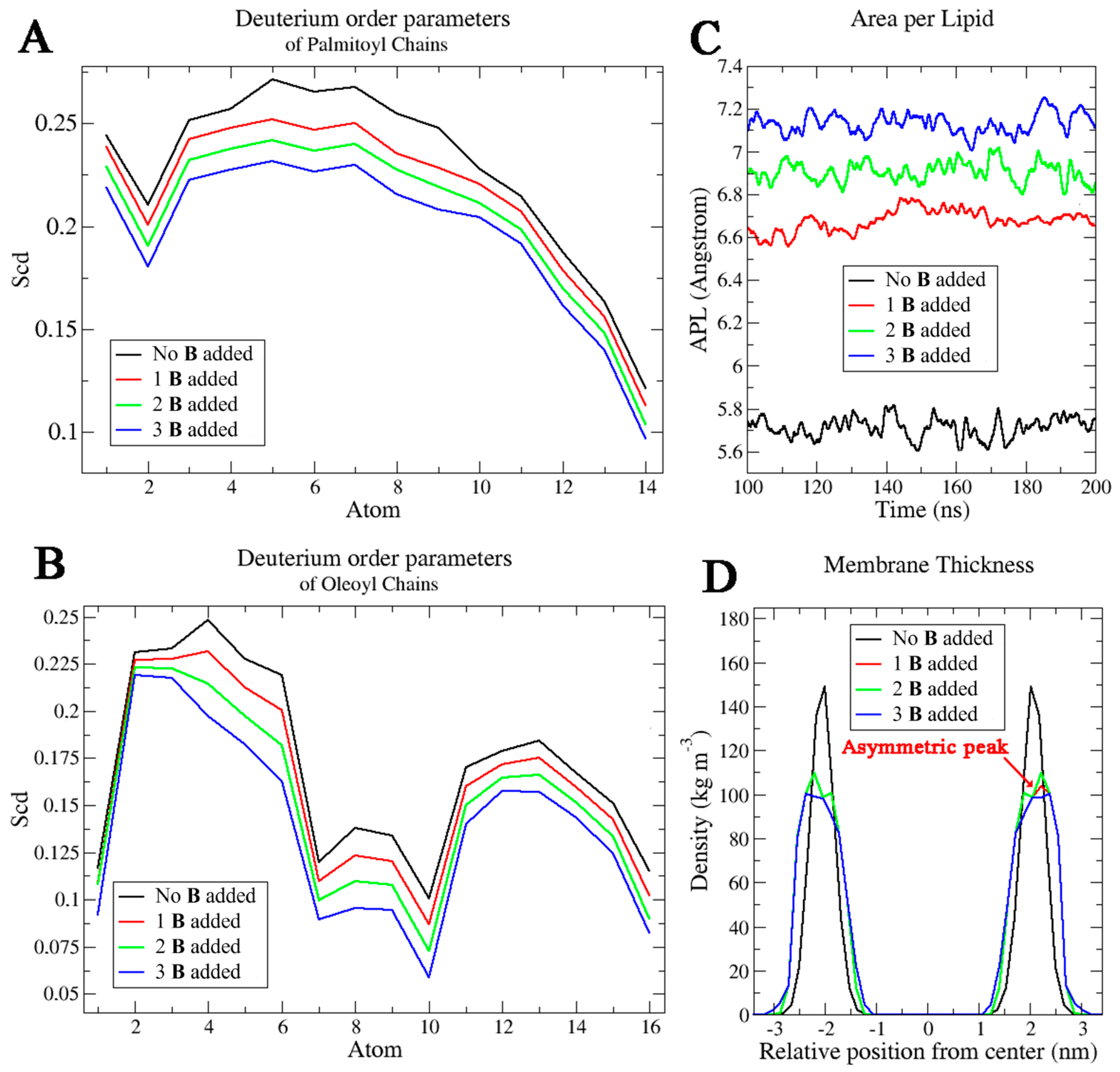
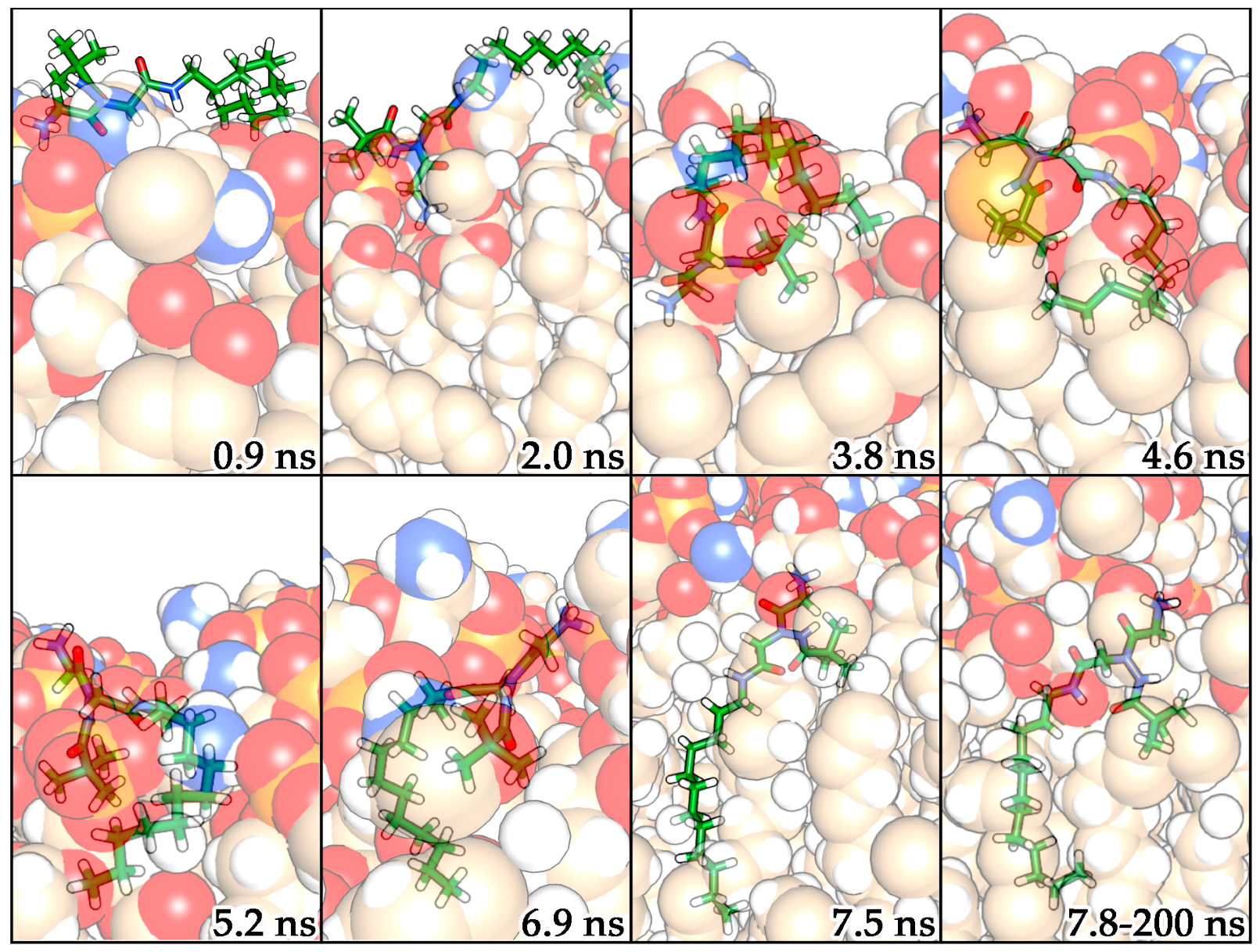
2.3. Molecular Dynamics Simulations
2.4. LPS Binding Assay
2.5. DNA Binding Assay
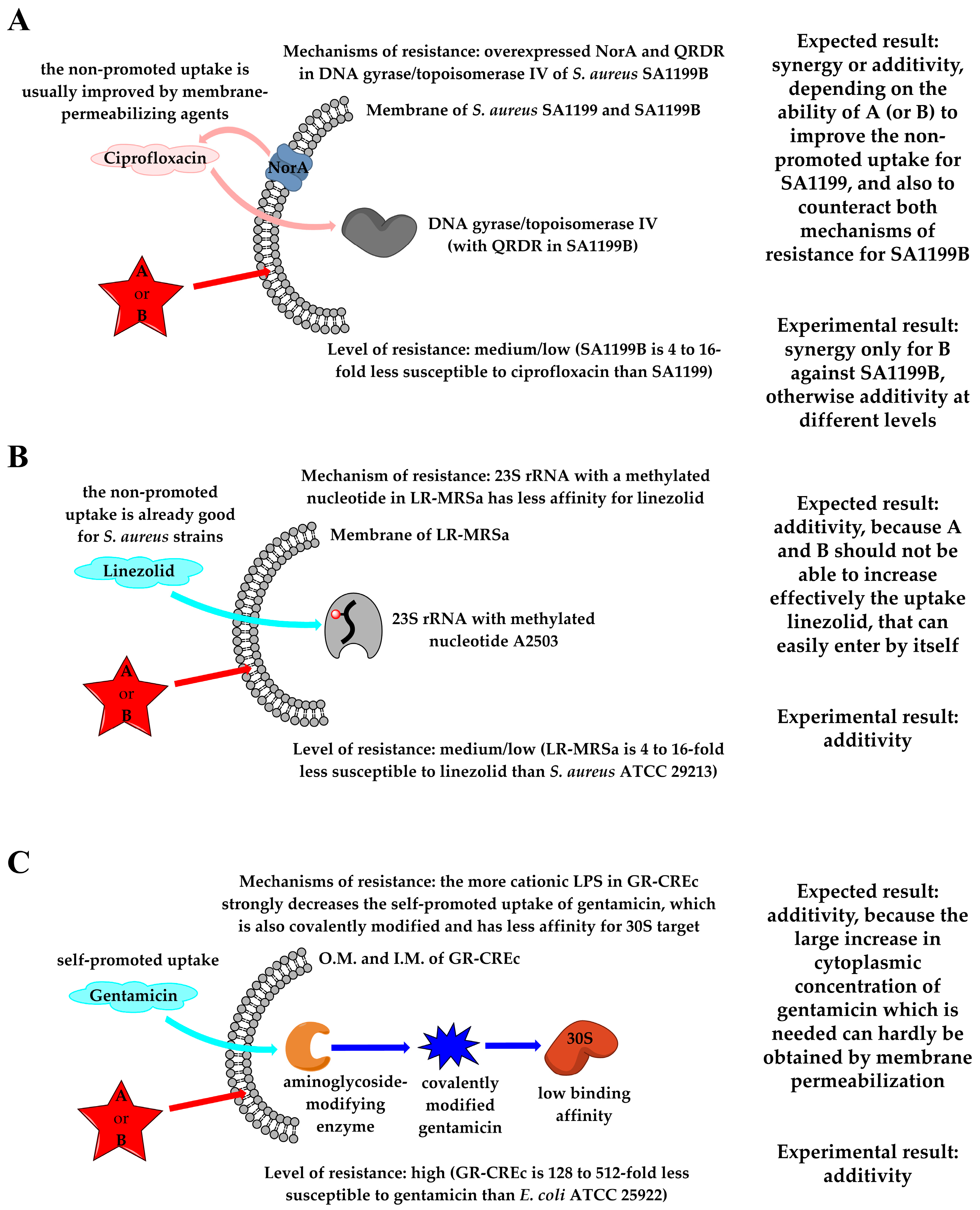
2.6. In Vitro Antimicrobial Susceptibility Assays
2.6.1. α-Hydrazido Acids Antimicrobial Activity
| MIC (μg/mL) a | ||||||
|---|---|---|---|---|---|---|
| S. aureus | E. coli | |||||
| Compd | ATCC 29213 | AOUC-0915 b | SA1199 c | SA1199B d | ATCC 25922 | 288328 e |
| A | 4 | 4 | 8 | 8 | 4 | 4 |
| B | 4 | 4 | 16 | 16 | 8 | 8 |
2.6.2. Combinations with First-Line Antibiotics
3. Materials and Methods
3.1. Density Functional Theory Calculations
3.2. MD Simulations
3.3. General Important Notes on the Use of α-Hydrazido Acid Hydrochlorides Solutions
3.4. Determination of CMC
3.5. DNA-Amphiphiles Binding Assay
3.6. LPS-Amphiphiles Binding Assay
3.7. Minimum Inhibitory Concentrations (MICs)
3.8. Checkerboard Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- See, for Example. Available online: https://www.who.int/antimicrobial-resistance/en/ (accessed on 27 March 2024).
- No Time to Wait: Securing the Future from Drug-Resistant Infections. 2019. Available online: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections (accessed on 25 March 2024).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance 2016. Wellcome Trust and HM Government. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 28 March 2024).
- Piddock, L.J.V. Reflecting on the final report of the O’Neill Review on Antimicrobial Resistance. Lancet Infect. Dis. 2016, 16, 767–768. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- McPhee, J.B.; Hancock, R.E.W. Function and therapeutic potential of host defense peptides. J. Pept. Sci. 2005, 11, 677–687. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K. Toxicity of polymyxins: A systematic review of the evidence from old and recent studies. Crit. Care 2006, 10, R27. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Gong, H.; Liao, M.; Hu, X.; Fa, K.; Phanphak, S.; Ciumac, D.; Hollowell, P.; Shen, K.; Clifton, L.A.; Campana, M.; et al. Aggregated Amphiphilic Antimicrobial Peptides Embedded in Bacterial Membranes. ACS Appl. Mater. Interfaces 2020, 12, 44420–44432. [Google Scholar] [CrossRef]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef]
- Liao, M.; Wu, C.; Shen, K.; Hu, X.; Lu, J.R. Combinatorial therapies of surfactant-like antimicrobial peptides and antibiotics for improved therapeutic efficacy. Curr. Opin. Colloid Interface Sci. 2024, 73, 101829. [Google Scholar] [CrossRef]
- Ejim, L.; Farha, M.A.; Falconer, S.B.; Wildenhain, J.; Coombes, B.K.; Tyers, M.; Brown, E.D.; Wright, G.D. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011, 7, 348–350. [Google Scholar] [CrossRef]
- Molchanova, N.; Hansen, P.R.; Franzyk, H. Advances in development of antimicrobial peptidomimetics as potential drugs. Molecules 2017, 22, 1430. [Google Scholar] [CrossRef] [PubMed]
- Raguse, T.L.; Porter, E.A.; Weisblum, B.; Gellman, S.H. Structure-Activity studies of 14-helical antimicrobial β-peptides: Probing the relationship between conformational stability and antimicrobial potency. J. Am. Chem. Soc. 2002, 124, 12774–12785. [Google Scholar] [CrossRef]
- Chongsiriwatana, N.P.; Miller, T.M.; Wetzler, M.; Vakulenko, S.; Karlsson, A.J.; Palecek, S.P.; Mobashery, S.; Barron, A.E. Short alkylated peptoid mimics of antimicrobial lipopeptides. Antimicrob. Agents Chemother. 2011, 55, 417–420. [Google Scholar] [CrossRef]
- Violette, A.; Fournel, S.; Lamour, K.; Chaloin, O.; Frisch, B.; Briand, J.P.; Monteil, H.; Guichard, G. Mimicking Helical Antibacterial Peptides with Nonpeptidic Folding Oligomers. Chem. Biol. 2006, 13, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Isaksson, J.; Brandsdal, B.O.; Engqvist, M.; Flaten, G.E.; Svendsen, J.S.M.; Stensen, W. A synthetic antimicrobial peptidomimetic (LTX 109): Stereochemical impact on membrane disruption. J. Med. Chem. 2011, 54, 5786–5795. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Isaacs, A.; Clements, D.; Liu, D.; Kim, H.; Scott, R.W.; Winkler, J.D.; Degrado, W.F. De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proc. Natl. Acad. Sci. USA 2009, 106, 6968–6973. [Google Scholar] [CrossRef]
- Thaker, H.D.; Cankaya, A.; Scott, R.W.; Tew, G.N. Role of amphiphilicity in the design of synthetic mimics of antimicrobial peptides with gram-negative activity. ACS Med. Chem. Lett. 2013, 4, 481–485. [Google Scholar] [CrossRef]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins). Biochim. Biophys. Acta Biomembr. 2007, 1768, 2500–2509. [Google Scholar] [CrossRef]
- Paulsen, M.H.; Engqvist, M.; Ausbacher, D.; Anderssen, T.; Langer, M.K.; Haug, T.; Morello, G.R.; Liikanen, L.E.; Blencke, H.M.; Isaksson, J.; et al. Amphipathic Barbiturates as Mimics of Antimicrobial Peptides and the Marine Natural Products Eusynstyelamides with Activity against Multi-resistant Clinical Isolates. J. Med. Chem. 2021, 64, 11395–11417. [Google Scholar] [CrossRef]
- Hoque, J.; Akkapeddi, P.; Yarlagadda, V.; Uppu, D.S.S.M.; Kumar, P.; Haldar, J. Cleavable cationic antibacterial amphiphiles: Synthesis, mechanism of action, and cytotoxicities. Langmuir 2012, 28, 12225–12234. [Google Scholar] [CrossRef]
- Hansen, T.; Last, T.; Havelkova, M.; Strøm, M.B. Antimicrobial activity of small β-peptidomimetics based on the pharmacophore model of short cationic antimicrobial peptides. J. Med. Chem. 2010, 53, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Bremner, J.B.; Keller, P.A.; Pyne, S.G.; Boyle, T.R.; Brkic, Z.; David, D.M.; Garas, A.; Morgan, J.; Robertson, M.; Somphol, K.; et al. Binaphthyl-based dicationic peptoids with therapeutic potential. Angew. Chem. Int. Ed. 2010, 49, 537–540. [Google Scholar] [CrossRef]
- Padhee, S.; Hu, Y.; Niu, Y.; Bai, G.; Wu, H.; Costanza, F.; West, L.; Harrington, L.; Shaw, L.N.; Cao, C.; et al. Non-hemolytic α-AApeptides as antimicrobial peptidomimetics. Chem. Commun. 2011, 47, 9729–9731. [Google Scholar] [CrossRef] [PubMed]
- Radzishevsky, I.S.; Rotem, S.; Bourdetsky, D.; Navon-Venezia, S.; Carmeli, Y.; Mor, A. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat. Biotechnol. 2007, 25, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Manjunath, G.B.; Akkapeddi, P.; Yarlagadda, V.; Hoque, J.; Uppu, D.S.S.M.; Konai, M.M.; Haldar, J. Small molecular antibacterial peptoid mimics: The simpler the better! J. Med. Chem. 2014, 57, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Mukherjee, S.; Patra, D.; Haldar, J. Polymeric Biomaterials for Prevention and Therapeutic Intervention of Microbial Infections. Biomacromolecules 2022, 23, 592–608. [Google Scholar] [CrossRef]
- Amabili, P.; Calvaresi, M.; Martelli, G.; Orena, M.; Rinaldi, S.; Sgolastra, F. Imidazolidinone-Tethered α-Hydrazidopeptides–Synthesis and Conformational Investigation. Eur. J. Org. Chem. 2019, 2019, 907–917. [Google Scholar] [CrossRef]
- Amabili, P.; Biavasco, F.; Brenciani, A.; Citterio, B.; Corbisiero, D.; Ferrazzano, L.; Fioriti, S.; Guerra, G.; Orena, M.; Rinaldi, S. Simple amphiphilic α-hydrazido acids: Rational design, synthesis, and in vitro bioactivity profile of a novel class of potential antimicrobial compounds. Eur. J. Med. Chem. 2020, 189, 112072. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmanna, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, F.; Chen, H.; Li, M.; Qiao, F.; Liu, Z.; Hou, Y.; Wu, C.; Fan, Y.; Liu, L.; et al. Selective Antimicrobial Activities and Action Mechanism of Micelles Self-Assembled by Cationic Oligomeric Surfactants. ACS Appl. Mater. Interfaces 2016, 8, 4242–4249. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Berglund, N.A.; Piggot, T.J.; Jefferies, D.; Sessions, R.B.; Bond, P.J.; Khalid, S. Interaction of the Antimicrobial Peptide Polymyxin B1 with Both Membranes of E. coli: A Molecular Dynamics Study. PLoS Comput. Biol. 2015, 11, e1004180. [Google Scholar] [CrossRef] [PubMed]
- Paracini, N.; Clifton, L.A.; Skoda, M.W.A.; Lakey, J.H. Liquid crystalline bacterial outer membranes are critical for antibiotic susceptibility. Proc. Natl. Acad. Sci. USA 2018, 115, E7587–E7594. [Google Scholar] [CrossRef]
- Mills, J.K.; Needham, D. Lysolipid incorporation in dipalmitoylphosphatidylcholine bilayer membranes enhances the ion permeability and drug release rates at the membrane phase transition. Biochim. Biophys. Acta Biomembr. 2005, 1716, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Róg, T.; Vattulainen, I. Cholesterol, sphingolipids, and glycolipids: What do we know about their role in raft-like membranes? Chem. Phys. Lipids 2014, 184, 82–104. [Google Scholar] [CrossRef]
- Shinoda, W. Permeability across lipid membranes. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2254–2265. [Google Scholar] [CrossRef]
- Sweet, M.J.; Hume, D.A. Endotoxin signal transduction in macrophages. J. Leukoc. Biol. 1996, 60, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Pulido, D.; Moussaoui, M.; Andreu, D.; Nogués, M.V.; Torrent, M.; Boix, E. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob. Agents Chemother. 2012, 56, 2378–2385. [Google Scholar] [CrossRef]
- Jerala, R.; Porro, M. Endotoxin Neutralizing Peptides. Curr. Top. Med. Chem. 2004, 4, 1173–1184. [Google Scholar] [CrossRef]
- Bhattacharjya, S. De novo Designed Lipopolysaccharide Binding Peptides: Structure Based Development of Antiendotoxic and Antimicrobial Drugs. Curr. Med. Chem. 2010, 17, 3080–3093. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Navarro, S.; Moussaoui, M.; Nogués, M.V.; Boix, E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 2008, 47, 3544–3555. [Google Scholar] [CrossRef]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of Action of the Antimicrobial Peptide Buforin II: Buforin II Kills Microorganisms by Penetrating the Cell Membrane and Inhibiting Cellular Functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef]
- Kosmidis, C.; Schindler, B.D.; Jacinto, P.L.; Patel, D.; Bains, K.; Seo, S.M.; Kaatz, G.W. Expression of multidrug resistance efflux pump genes in clinical and environmental isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents 2012, 40, 204–209. [Google Scholar] [CrossRef]
- Felicetti, T.; Mangiaterra, G.; Cannalire, R.; Cedraro, N.; Pietrella, D.; Astolfi, A.; Massari, S.; Tabarrini, O.; Manfroni, G.; Barreca, M.L.; et al. C-2 phenyl replacements to obtain potent quinoline-based Staphylococcus aureus NorA inhibitors. J. Enzym. Inhib. Med. Chem. 2020, 35, 584–597. [Google Scholar] [CrossRef]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Terminology relating to methods for the determination of susceptibility of bacteria to antimicrobial agents. Clin. Microbiol. Infect. 2000, 6, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Meletiadis, J.; Pournaras, S.; Roilides, E.; Walsh, T.J. Defining fractional inhibitory concentration index cutoffs for additive interactions based on self-drug additive combinations, Monte Carlo simulation analysis, and in vitro-in vivo correlation data for antifungal drug combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2010, 54, 602–609. [Google Scholar]
- Choi, H.; Lee, D.G. Synergistic effect of antimicrobial peptide arenicin-1 in combination with antibiotics against pathogenic bacteria. Res. Microbiol. 2012, 163, 479–486. [Google Scholar] [CrossRef]
- Fratini, F.; Mancini, S.; Turchi, B.; Friscia, E.; Pistelli, L.; Giusti, G.; Cerri, D. A novel interpretation of the Fractional Inhibitory Concentration Index: The case Origanum vulgare L. and Leptospermum scoparium J. R. et G. Forst essential oils against Staphylococcus aureus strains. Microbiol. Res. 2017, 195, 11–17. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta-Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Chongsiriwatana, N.P.; Wetzler, M.; Barron, A.E. Functional synergy between antimicrobial peptoids and peptides against gram-negative bacteria. Antimicrob. Agents Chemother. 2011, 55, 5399–5402. [Google Scholar] [CrossRef]
- Chukwudi, C.U. rRNA binding sites and the molecular mechanism of action of the tetracyclines. Antimicrob. Agents Chemother. 2016, 60, 4433–4441. [Google Scholar] [CrossRef]
- Antonelli, A.; D’Andrea, M.M.; Galano, A.; Borchi, B.; Brenciani, A.; Vaggelli, G.; Cavallo, A.; Bartoloni, A.; Giovanetti, E.; Rossolini, G.M. Linezolid-resistant cfr-positive MRSA, Italy. J. Antimicrob. Chemother. 2016, 71, 2349–2351. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, A.; Tariq, A.; Iqbal, M.; Mirza, O.; Haque, A.; Walz, T.; Rahman, M. Mutational Diversity in the Quinolone Resistance-Determining Regions of Type-II Topoisomerases of Salmonella Serovars. Antibiotics 2021, 10, 1455. [Google Scholar] [CrossRef]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739. [Google Scholar] [CrossRef] [PubMed]
- Simoni, S.; Morroni, G.; Brenciani, A.; Vincenzi, C.; Cirioni, O.; Castelletti, S.; Varaldo, P.E.; Giovanetti, E.; Mingoia, M. Spread of colistin resistance gene mcr-1 in Italy: Characterization of the mcr-1.2 allelic variant in a colistin-resistant blood isolate of Escherichia coli. Diagn. Microbiol. Infect. Dis. 2018, 91, 66–68. [Google Scholar] [CrossRef]
- Castanheira, M.; Davis, A.P.; Serio, A.W.; Krause, K.M.; Mendes, R.E. In vitro activity of Plazomicin against Enterobacteriaceae isolates carrying genes encoding aminoglycoside-modifying enzymes most common in US Census divisions. Diagn. Microbiol. Infect. Dis. 2019, 94, 73–77. [Google Scholar] [CrossRef]
- Cirioni, O.; Simonetti, O.; Pierpaoli, E.; Barucca, A.; Ghiselli, R.; Orlando, F.; Pelloni, M.; Minardi, D.; Trombettoni, M.M.C.; Guerrieri, M.; et al. Enhanced efficacy of combinations of pexiganan with colistin versus Acinetobacter baumannii in experimental sepsis. Shock 2016, 46, 219–225. [Google Scholar] [CrossRef]
- Morroni, G.; Simonetti, O.; Brenciani, A.; Brescini, L.; Kamysz, W.; Kamysz, E.; Neubauer, D.; Caffarini, M.; Orciani, M.; Giovanetti, E.; et al. In vitro activity of Protegrin-1, alone and in combination with clinically useful antibiotics, against Acinetobacter baumannii strains isolated from surgical wounds. Med. Microbiol. Immunol. 2019, 208, 877–883. [Google Scholar] [CrossRef]
- Scott, M.G.; Yan, H.; Hancock, R.E.W. Biological Properties of Structurally Related-Helical Cationic Antimicrobial Peptides. Infect. Immun. 1999, 67, 2005–2009. [Google Scholar] [CrossRef]
- Wu, X.; Li, Z.; Li, X.; Tian, Y.; Fan, Y.; Yu, C.; Zhou, B.; Liu, Y.; Xiang, R.; Yang, L. Synergistic effects of antimicrobial peptide DP7 combined with antibiotics against multidrug-resistant bacteria. Drug Des. Devel. Ther. 2017, 11, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Arenas, I.; Villegas, E.; Walls, O.; Barrios, H.; Rodríguez, R.; Corzo, G.; Rutledge, P.J. Antimicrobial activity and stability of short and long based arachnid synthetic peptides in the presence of commercial antibiotics. Molecules 2016, 21, 225. [Google Scholar] [CrossRef] [PubMed]
- Kampshoff, F.; Willcox, M.D.P.; Dutta, D. A pilot study of the synergy between two antimicrobial peptides and two common antibiotics. Antibiotics 2019, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Soren, O.; Brinch, K.S.; Patel, D.; Liu, Y.; Liu, A.; Coates, A.; Hu, Y. Antimicrobial peptide novicidin synergizes with rifampin, ceftriaxone, and ceftazidime against antibiotic-resistant Enterobacteriaceae in vitro. Antimicrob. Agents Chemother. 2015, 59, 6233–6240. [Google Scholar] [CrossRef]
- Giacometti, A.; Cirioni, O.; Ghiselli, R.; Mocchegiani, F.; Orlando, F.; Silvestri, C.; Bozzi, A.; Di Giulio, A.; Luzi, C.; Mangoni, M.L.; et al. Interaction of antimicrobial peptide temporin L with lipopolysaccharide in vitro and in experimental rat models of septic shock caused by gram-negative bacterial. Antimicrob. Agents Chemother. 2006, 50, 2478–2486. [Google Scholar] [CrossRef]
- Marcellini, L.; Borro, M.; Gentile, G.; Rinaldi, A.C.; Stella, L.; Aimola, P.; Barra, D.; Mangoni, M.L. Esculentin-1b(1-18)—A membrane-active antimicrobial peptide that synergizes with antibiotics and modifies the expression level of a limited number of proteins in Escherichia coli. FEBS J. 2009, 276, 5647–5664. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Abdelkhalek, A.; Seleem, M.N. Evaluation of short synthetic antimicrobial peptides for treatment of drug-resistant and intracellular Staphylococcus aureus. Sci. Rep. 2016, 6, 29707. [Google Scholar] [CrossRef]
- Munita, J.M.; Bayer, A.S.; Arias, C.A. Evolving Resistance among Gram-positive Pathogens. Clin. Inf. Dis. 2015, 61, S48–S57. [Google Scholar] [CrossRef]
- Fishovitz, J.; Hermoso, J.A.; Chang, M.; Mobashery, S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 2014, 66, 572–577. [Google Scholar] [CrossRef]
- Shah, P.; Chen, C.S. Combinational Synergy of Antibiotics and Antimicrobial Peptides. J. Transl. Proteomics Res. 2017, 3, 6–11. [Google Scholar]
- Duong, L.; Gross, S.P.; Siryaporn, A. Developing Antimicrobial Synergy with AMPs. Front. Med. Technol. 2021, 3, 640981. [Google Scholar] [CrossRef] [PubMed]
- Oo, T.Z.; Cole, N.; Garthwaite, L.; Willcox, M.D.P.; Zhu, H. Evaluation of synergistic activity of bovine lactoferricin with antibiotics in corneal infection. J. Antimicrob. Chemother. 2010, 65, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Long, K.S.; Vester, B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 2012, 56, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Farmer, S.W.; Li, Z.; Poolet, K. Interaction of Aminoglycosides with the Outer Membranes and Purified Lipopolysaccharide and OmpF Porin of Escherichia coli. Antimicrob. Agents Chemother. 1991, 35, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.M.; Ortega, M.; Tarragó, C.; Albet, C.; Vila, J.; Terencio, J.; Guglietta, A. Decreased linezolid uptake in an in vitro-selected linezolid-resistant Staphylococcus epidermidis mutant. J. Antimicrob. Chemother. 2009, 64, 990–992. [Google Scholar] [CrossRef]
- Peterson, A.A.; Fesik, S.W.; McGroarty, E.J. Decreased Binding of Antibiotics to Lipopolysaccharides from Polymyxin-Resistant Strains of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 1987, 31, 230–237. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Da Chai, J.; Head-Gordon, M. Systematic optimization of long-range corrected hybrid density functionals. J. Chem. Phys. 2008, 128, 084106. [Google Scholar] [CrossRef]
- Lin, Y.S.; De Li, G.; Mao, S.P.; Da Chai, J. Long-range corrected hybrid density functionals with improved dispersion corrections. J. Chem. Theory Comput. 2013, 9, 263–272. [Google Scholar] [CrossRef]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum mechanical continuum solvation models. Chem. Rev. 2005, 105, 2999–3093. [Google Scholar] [CrossRef]
- Park, S.J.; Kern, N.; Brown, T.; Lee, J.; Im, W. CHARMM-GUI PDB Manipulator: Various PDB Structural Modifications for Biomolecular Modeling and Simulation. J. Mol. Biol. 2023, 435, 167995. [Google Scholar] [CrossRef]
- Pandit, K.R.; Klauda, J.B. Membrane models of E. coli containing cyclic moieties in the aliphatic lipid chain. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1205–1210. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Páll, S.; Hess, B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1997, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Tsutsumi-Yasuhara, S.; Ookoshi, T.; Ohhashi, Y.; Kimura, H.; Takahashi, N.; Yoshida, H.; Miyazaki, R.; Goto, Y.; Naiki, H. Growth of β2-microglobulin-related amyloid fibrils by non-esterified fatty acids at a neutral pH. Biochem. J. 2008, 416, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef]
- Wood, S.J.; Miller, K.A.; David, S.A. Anti-Endotoxin Agents. 1. Development of a Fluorescent Probe Displacement Method Optimized for High-Throughput Identification of Lipopolysaccharide-Binding Agents. Comb. Chem. High Throughput Screen. 2004, 7, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.B.; Cockerill, F.R., III; Bradford, P.A.; Eliopoulos, G.M.; Hindler, J.A.; Jenkins, S.G.; Lewis, J.S., II; Limbago, B.; Miller, L.A.; Nicolau, D.P.; et al. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Tenth Edition (2017) Document M07-A10, Volume 35, No 2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]


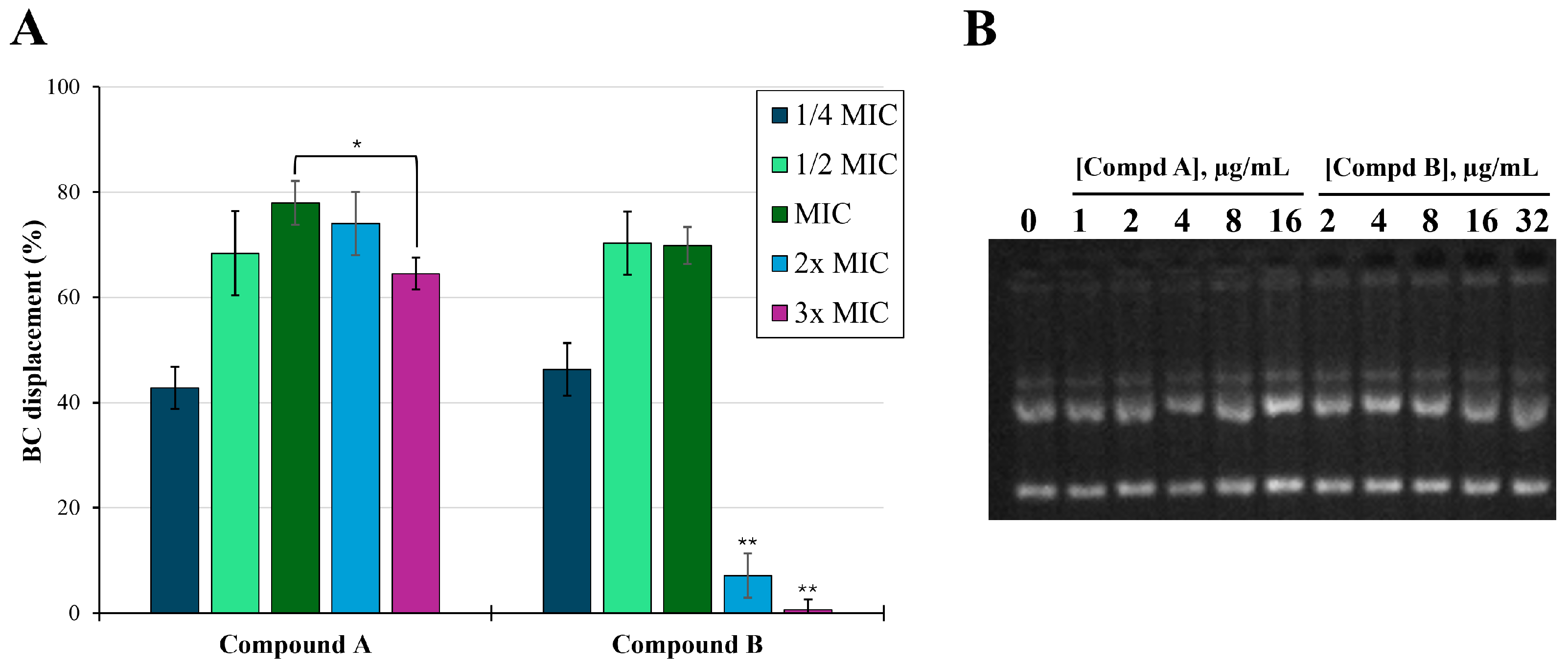

| ΣFIC vs. Drug Sensitive Bacteria ([H]/[X]) b for Combinations with the Indicated Antibiotics | |||||
| Compd | S. aureus ATCC 29213 c | S. aureus SA1199 d | E. coli ATCC 25922 c | ||
| Tetracycline | Ciprofloxacin | Tetracycline | |||
| A | 0.50 (1/0.0313) | 2 (8/0.16) | 0.38 (1/0.0625) | ||
| B | 0.38 (1/0.0156) | 0.75 (4/0.08) | 0.50 (2/0.125) | ||
| ΣFIC vs. Drug Resistant Bacteria ([H]/[X]) b for Combinations with the Indicated Antibiotics | |||||
| S. aureus AOUC-0915 (LR-MRSa) e | S. aureus SA1199B f | E. coli 288328 (GR-CREc) g | |||
| Methicillin h | Linezolid | Ciprofloxacin | Colistin | Gentamicin | |
| A | 0.5 < ΣFIC ≤ 0.53 (2/64) | 1.03 (0.125/16) | 0.75 (4/2.5) | 0.31 (1/0.5) | 1.25 (1/128) |
| B | 0.5 < ΣFIC ≤ 1.0 (2/1024) | 1.03 (0.125/16) | 0.5 (4/2.5) | 0.28 (2/0.25) | 1.50 (4/128) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minnelli, C.; Mangiaterra, G.; Laudadio, E.; Citterio, B.; Rinaldi, S. Investigation on the Synergy between Membrane Permeabilizing Amphiphilic α-Hydrazido Acids and Commonly Used Antibiotics against Drug-Resistant Bacteria. Molecules 2024, 29, 4078. https://doi.org/10.3390/molecules29174078
Minnelli C, Mangiaterra G, Laudadio E, Citterio B, Rinaldi S. Investigation on the Synergy between Membrane Permeabilizing Amphiphilic α-Hydrazido Acids and Commonly Used Antibiotics against Drug-Resistant Bacteria. Molecules. 2024; 29(17):4078. https://doi.org/10.3390/molecules29174078
Chicago/Turabian StyleMinnelli, Cristina, Gianmarco Mangiaterra, Emiliano Laudadio, Barbara Citterio, and Samuele Rinaldi. 2024. "Investigation on the Synergy between Membrane Permeabilizing Amphiphilic α-Hydrazido Acids and Commonly Used Antibiotics against Drug-Resistant Bacteria" Molecules 29, no. 17: 4078. https://doi.org/10.3390/molecules29174078







