Discovery of Coumarins from Zanthoxylum dimorphophyllum var. spinifoliumas and Their Potential against Rheumatoid Arthritis
Abstract
:1. Introduction
2. Results
2.1. Structural Elucidation of Eight New Compounds
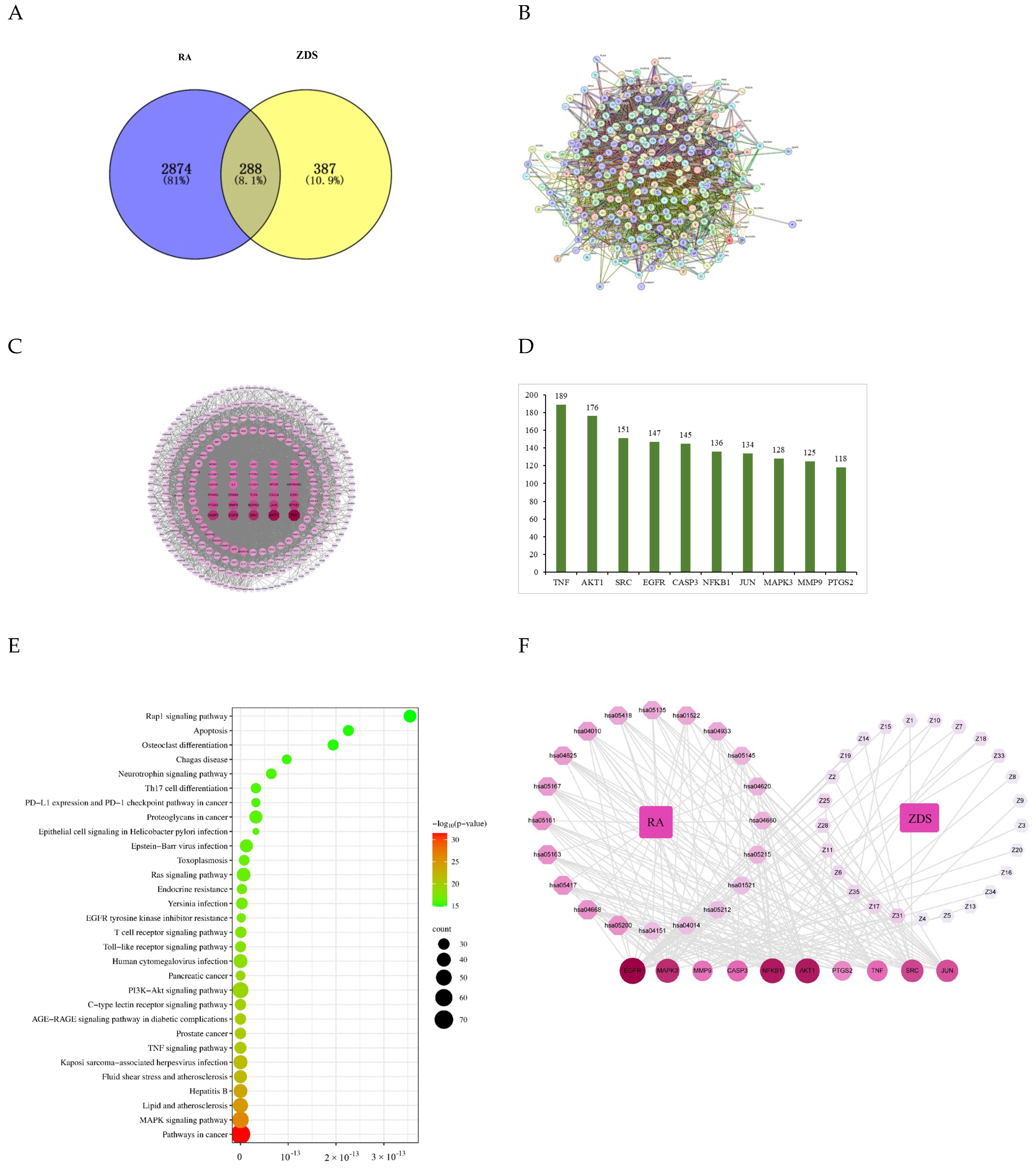
2.2. Network Pharmacology
2.2.1. Construction and Analysis of the PPI Network
2.2.2. KEGG Enrichment Analysis
2.2.3. Compound-Target-Pathway Network Analyses
2.3. Experimental Validation of the Anti-RA Action of ZDS In Vitro
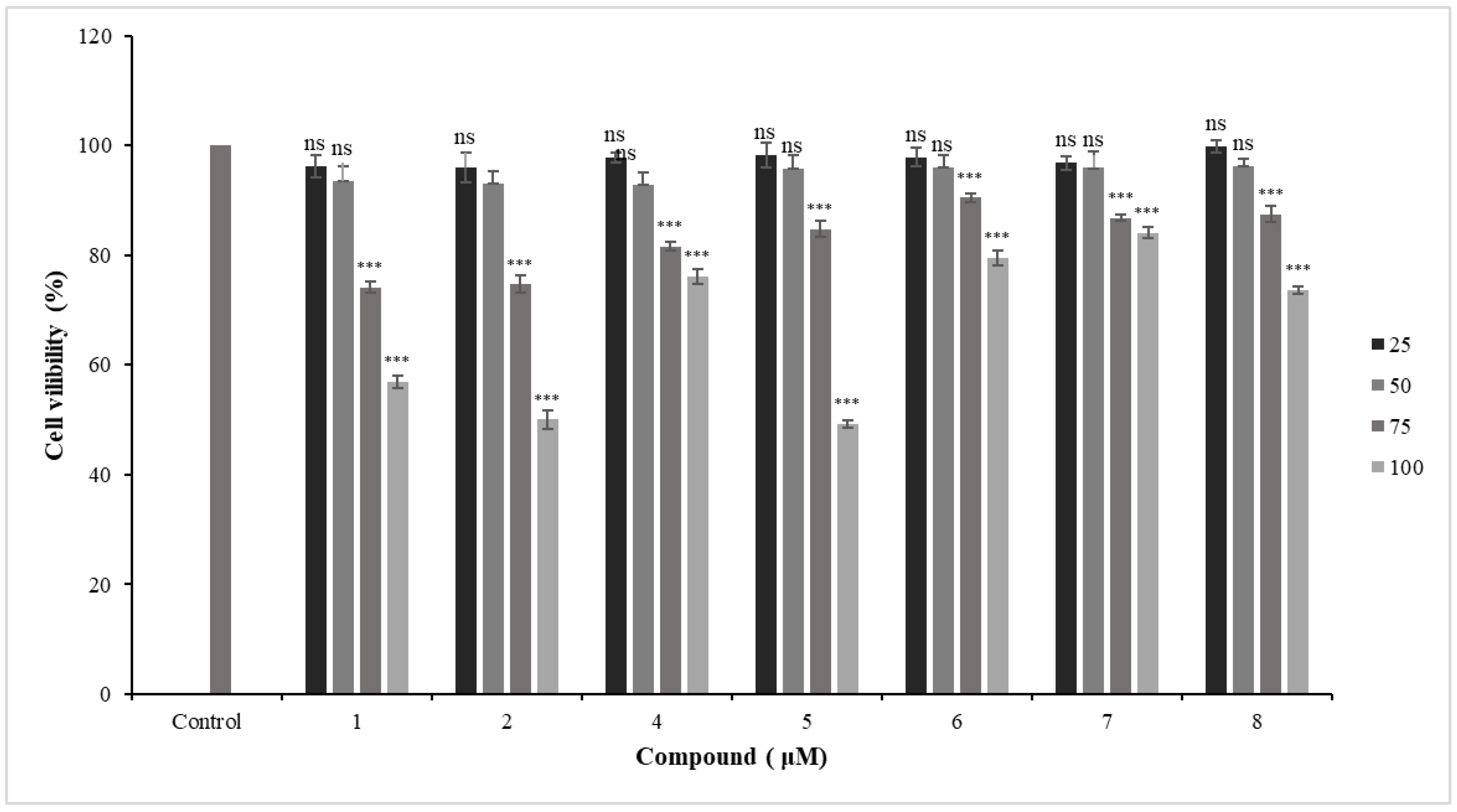
2.3.1. Cytotoxic Effects of New Coumarins in Primary HFLS-RA Cells
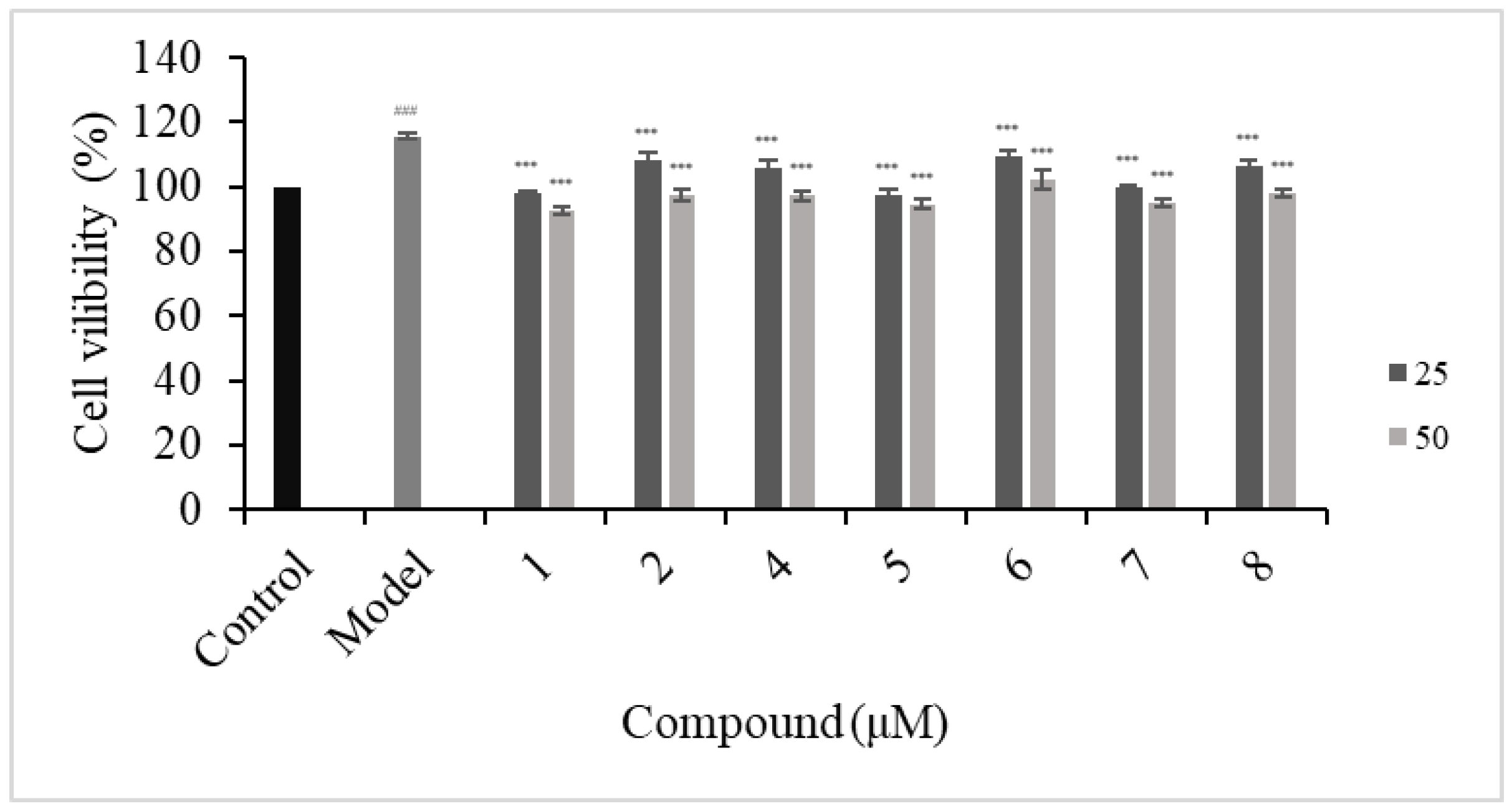
2.3.2. Antiproliferative of HFLS-RA Cells
2.3.3. Effects of Compounds 1, 5, and 7 on IL-1β, IL-6, and TNF-α Secretions
2.4. Molecular Docking Analysis
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material
4.3. Extract and Isolation
4.4. Electronic Circular Dichroism Calculation of Compounds 7 and 8
4.5. Determination of the Absolute Configuration of 1 and 7
4.5.1. Preparation of the Rh2(OCOCF3)4 Complex of Compound 1
4.5.2. Preparation of the Mo2(OAc)4 Complex of Compound 7
4.6. Network Pharmacology Analysis
4.6.1. Target Prediction of ZDS
4.6.2. Acquisition of RA-Associated Genes
4.6.3. Construction of a Protein–Protein Interaction Network
4.6.4. KEGG Analysis
4.6.5. Network Construction and Analysis
4.7. Anti-RA Activity Screening
4.7.1. Cell Culture
4.7.2. Cell Cytotoxic Assay
4.7.3. Cell Proliferation Assay
4.7.4. Enzyme-Linked Immunosorbent Assay (ELISA)
4.7.5. Statistical Analysis
4.8. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meng, F.X.; Tao, X.M. Effect of key enzyme inhibition of glycolysis on synovial fibroblasts in Rheumatoid Arthritis. J. Biol. Pharm. Open 2021, 2, 20–26. [Google Scholar] [CrossRef]
- Slobodin, G.; Shoenfeld, Y. Rheumatic Disease in Geriatrics: Diagnosis and Management; Springer Nature: Cham, Switzerland, 2020; p. 173. [Google Scholar] [CrossRef]
- GBD 2021 Rheumatoid Arthritis Collaborators. Global regional, and national burden of rheumatoid arthritis, 1990–2020, and projections to 2050: A systematic analysis of the Global Burden of Disease Study 2021. Lancet Rheumatol. 2023, 5, e594–e610. [Google Scholar] [CrossRef] [PubMed]
- Schett, G.; Gravallese, E. Bone erosion in rheumatoid arthritis: Mechanisms, diagnosis and treatment. Nat. Rev. Rheumatol. 2012, 8, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Bugatti, S.; Codullo, V.; Caporali, R.; Montecucco, C. B cells in rheumatoid arthritis. Autoimmun. Rev. 2007, 7, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Marston, B.; Palanichamy, A.; Anolik, J.H. B cells in the pathogenesis and treatment of rheumatoid arthritis. Curr. Opin. Rheumatol. 2010, 22, 307–315. [Google Scholar] [CrossRef]
- Noss, E.H.; Brenner, M.B. The role and therapeutic implications of fibroblast-like synoviocytes in inflammation and cartilage erosion in rheumatoid arthritis. Immunol. Rev. 2008, 223, 252–270. [Google Scholar] [CrossRef]
- Miao, Y.H.; Yang, J.; Yun, Y.L.; Sun, J.; Wang, X.J. Synthesis and anti-rheumatoid arthritis activities of 3-(4-aminophenyl)-coumarin derivatives. J. Enzym. Inhib. Med. Chem. 2021, 36, 450–461. [Google Scholar] [CrossRef]
- Zhang, W.J.; Guan, W.; Geng, Z.F.; Wang, Y.; Pang, X.; You, C.X.; Du, S.S. Two new coumarins from Zanthoxylum dimorphophyllum spinifolium and their feeding deterrent activities against Tribolium castaneum. Ind. Crop. Prod. 2020, 143, 111889. [Google Scholar] [CrossRef]
- Zhou, X.J.; Gao, Y.X.; Hu, L.P.; Xie, T.Z.; Zhang, J. Analysis of leaf oil of Zanthoxylum ovalifolium var. spinifolium Rehd. Et. Wils by GC-MS. Res. Dev. Mark. 2009, 25, 490–491. [Google Scholar]
- Tao, Z.Y.; Chen, W.S.; Zhang, W.D.; Zheng, S.Q.; Qiao, C.Z. Studies on the coumarins in the root of Zanthoxylum dimorphophyllum. Chin. J. Chin. Mat. Med. 2003, 28, 344–346. [Google Scholar]
- Xiang, Y.; Zheng, Q.A.; Zhang, C.K.; Li, Z.; Tu, Z.B. Alkaloids and coumarins from Zanthoxylum dimorphophyllum Hemsl. var. spinifolium Rehd ET Wils. J. Wuhan Bot. Res. 2000, 18, 143–145. [Google Scholar]
- Tao, Z.Y.; Chen, W.S.; Zhang, W.D.; Zheng, S.Q.; Qiao, C.Z. Lignans from the roots of Zanthoxylum dimorphophyllum spinifolium. Chin. Tradit. Herb. Drugs 2004, 4, 378–379. [Google Scholar]
- Gangopadhyay, A. Plant-derived natural coumarins with anticancer potentials: Future and challenges. J. Herb. Med. 2023, 42, 100797. [Google Scholar] [CrossRef]
- Liang, W.Y.; Yang, H.Y.; Lei, H.X.; Xiang, Z.B.; Duan, Y.Q.; Xin, H.L.; Han, T.; Su, J. Phytochemistry and health functions of Zanthoxylum bungeanum Maxim and Zanthoxylum schinifolium Sieb. et Zucc as pharma-foods: A systematic review. Trends Food Sci. Technol. 2024, 143, 104225. [Google Scholar] [CrossRef]
- Li, M.L.; Lei, P.; Shuang, S.M.; Dong, C.; Zhang, L.Y. Visualization of polarity changes in endoplasmic reticulum (ER) autophagy and rheumatoid arthritis mice with near-infrared ER-targeted fluorescent probe. Talanta 2024, 275, 126141. [Google Scholar] [CrossRef]
- Pan, R.; Gao, X.H.; Li, Y.; Xia, Y.F.; Dai, Y. Anti-arthritic effect of scopoletin, a coumarin compound occurring in Erycibe obtusifolia Benth stems, is associated with decreased angiogenesis in synovium. Fundam. Clin. Pharmacol. 2010, 24, 477–490. [Google Scholar] [CrossRef]
- Emanuela, B.; Laura, M.; Simone, C.; Lorenzo, C.M.; Paolo, G.; Alessandro, D.L.; Fabrizio, C.; Carla, G.; Daniela, S.; Claudiu, T.S. Novel Insights on CAI−CORM Hybrids: Evaluation of the CO Releasing Properties and Pain-Relieving Activity of Differently Substituted Coumarins for the Treatment of Rheumatoid Arthritis. J. Med. Chem. 2023, 66, 1892–1908. [Google Scholar] [CrossRef]
- Dorababu, A. Pharmacological report of recently designed multifunctional coumarin and coumarin-heterocycle derivatives. Arch. Pharm. 2022, 355, e2100345. [Google Scholar] [CrossRef]
- Cho, J.Y.; Hwang, T.L.; Chang, T.H.; Lim, Y.P.; Sung, P.J.; Lee, T.H.; Chen, J.J. New coumarins and anti-inflammatory constituents from Zanthoxylum avicennae. Food Chem. 2012, 135, 17–23. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Zhao, B.T.; Kim, O.; Lee, J.H.; Choi, J.S.; Min, B.S.; Woo, M.H. Anti-inflammatory terpenylated coumarins from the leaves of Zanthoxylum schinifolium with α-glucosidase inhibitory activity. J. Nat. Med. 2016, 70, 276–281. [Google Scholar] [CrossRef]
- Frelek, J.; Szczepek, W.J. [Rh2(OCOCF3)4] as an auxiliary chromophore in chiroptical studies on steroidal alcohols. Tetrahedron Asymmetry 1999, 10, 1507–1520. [Google Scholar] [CrossRef]
- Gorecki, M.; Jablonska, E.; Kruszewska, A.; Suszczynska, A.; Urbanczyk-Lipkowska, Z.; Gerards, M.; Morzycki, J.W.; Szczepek, W.J.; Frelek, J. Practical method for the absolute configuration assignment of tert/tert 1,2-diols using their complexes with Mo2(OAc)4. J. Org. Chem. 2007, 72, 2906–2916. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.V.; Rao, K.S.; Annamalai, T.; Mukhopadhyay, T. New coumarin diol from the plant, Chloroxylon swietenia DC. Indian J. Chem. 2009, 48, 1041–1044. [Google Scholar]
- Kwon, O.S.; Choi, J.S.; Islam, M.N.; Kim, Y.S.; Kim, H.P. Inhibition of 5-lipoxygenase and skin inflammation by the aerial parts of Artemisia capillaris and its constituents. Arch. Pharm. Res. 2011, 34, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.P.; Yan, G.; Guo, J.M.; Liu, Y.Y.; Li, Y.J.; Zhao, Y.Y.; Qiang, L.; Fu, Y.H. Prenylated coumarins from the fruits of Manilkara zapota with potential anti-inflammatory effects and anti-HIV activities. J. Agric. Food. Chem. 2019, 67, 11942–11947. [Google Scholar] [CrossRef] [PubMed]
- Farokhi-Firoozi, H.; Rahavi, M.; Pirali-Hamedani, M.; Hadjiakhoondi, A.; Delnavazi, M. Essential oil analysis and isolation of coumarins and flavonol glycosides of Ferulago angulata (Schltdl.) Boiss. Fruits. J. Pharm. Sci. 2021, 27, 139–146. [Google Scholar] [CrossRef]
- Park, J.S.; Chung, B.; Lee, W.H.; Lee, J.; Suh, Y.; Oh, D.C.; Oh, K.B.; Shin, J. Sortase a-inhibitory coumarins from the folk medicinal plant Poncirus trifoliata. J. Nat. Prod. 2020, 83, 3004–3011. [Google Scholar] [CrossRef]
- Nayeli, M.B.; Maribel, H.R.; Enrique, J.F.; Rafael, B.P.; Margarita, A.F.; Macrina, F.M.; Ivan, M.D.; Manasés, G.C. Anti-inflammatory activity of coumarins isolated from Tagetes lucida Cav. Nat. Prod. Res. 2020, 34, 3244–3248. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Yang, X.W.; Wu, B.F.; Shang, J.H.; Luo, X.D. Anti-inflammatory effect of Pomelo peel and its bioactive coumarins. J. Agric. Food Chem. 2019, 67, 8810–8818. [Google Scholar] [CrossRef]
- He, R.J.; Huang, X.S.; Zhang, Y.J.; Wu, L.D.; Nie, H.; Zhou, D.X.; Liu, B.M.; Deng, S.P.; Yang, R.Y.; Huang, S.A.; et al. Structural characterization and assessment of the cytotoxicity of 2,3-dihydro-1-H-indene derivatives and coumarin glucosides from the bark of Streblus indicus. J. Nat. Prod. 2016, 79, 2472–2478. [Google Scholar] [CrossRef]
- Bissoue, A.N.; Muyard, F.; Bevalot, F.; Tillequin, F.; Mercier, M.F.; Armstrong, J.A.; Vaquette, J.; Waterman, P.G. Coumarins from the aerial parts of Chorilaena quercifolia. Phytochemistry 1996, 43, 877–879. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Li, W.; Yang, X.W. Biotransformation of columbianadin by rat hepatic microsomes and inhibition of biotransformation products on NO production in RAW 264.7 cells in vitro. Phytochemistry 2012, 81, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Takasugi, M.; Anetai, M. Psoralen and other linear furanocoumarins as phytoalexins in Glehnia littoralis. Phytochemistry 1998, 47, 13–16. [Google Scholar] [CrossRef]
- Lee, B.W.; Ha, T.K.Q.; Cho, H.M.; An, J.P.; Kim, S.K.; Kim, C.S.; Kimc, E.; Oh, W.K. Antiviral activity of furanocoumarins isolated from Angelica dahurica against influenza a viruses H1N1 and H9N2. J. Ethnopharmacol. 2020, 259, 112945. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wang, M.; Gao, M.; Zhao, M.; Bai, Y.; Zhao, C. Chemical composition and biological activities of Gerbera anandria. Molecules 2014, 19, 4046–4057. [Google Scholar] [CrossRef]
- Karakaya, S.; Şimşek, D.; Özbek, H.; Güvenalp, Z.; Altanlarb, N.; Kiliç, C.S. Antimicrobial activities of extracts and isolated coumarins from the roots of Four Ferulago species growing in Turkey. Iran. J. Pharm. Res. 2019, 18, 1516–1529. [Google Scholar] [CrossRef]
- Kim, Y.A.; Kong, C.S.; Park, H.H.; Lee, E.; Jang, M.S.; Nam, K.H.; Seo, Y. Anti-inflammatory activity of heterocarpin from the Salt marsh plant corydalis heterocarpa in LPS-induced RAW 264.7 Macrophage Cells. Molecules 2015, 20, 14474–14486. [Google Scholar] [CrossRef]
- Bai, J.R.; Xie, N.; Hou, Y.; Chen, X.R.; Hu, Y.; Zhang, Y.; Meng, X.L.; Wang, X.B.; Tang, C. The enhanced mitochondrial dysfunction by cantleyoside confines inflammatory response and promotes apoptosis of human HFLS-RA cell line via AMPK/Sirt 1/NF-κB pathway activation. Biomed. Pharmacother. 2022, 149, 112847. [Google Scholar] [CrossRef]
- Trid, S.; Ratchuporn, S.; Nadchawan, C.; Rose, S.S. Zanthoxylum spp.: A new potential sources of essential oil for the perfumery and pharmaceutical industries in Thailand. Med. Plants Int. J. Phytomed. Relat. Ind. 2019, 11, 26–45. [Google Scholar] [CrossRef]
- Mutinda, E.S.; Kimutai, F.; Mkala, E.M.; Waswa, E.N.; Odago, W.O.; Nanjala, C.; Ndungu, C.N.; Gichua, M.K.; Njire, M.M.; Gituru, R.W.; et al. Ethnobotanical uses, phytochemistry and pharmacology of pantropical genus Zanthoxylum L. (Rutaceae): An update. J. Ethnopharmacol. 2023, 303, 115895. [Google Scholar] [CrossRef]
- Sashidhara, K.V.; Kumar, M.; Modukuri, R.K.; Sonkar, R.; Bhatia, G.; Khanna, A.K.; Rai, S.; Shukla, R. Synthesis and Anti-inflammatory activity of novel biscoumarin–Chalcone hybrids. Bioorg. Med. Chem. Lett. 2011, 21, 4480–4484. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Song, X.; Lin, D.; Xu, P. Isofraxidin alleviates myocardial infarction through NLRP3 inflammasome inhibition. Inflammation 2020, 43, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Espírito-Santo, R.F.; Meira, C.S.; Costa, R.; Dos, S.; Filho, O.P.S.; Evangelista, A.F.; Trossini, G.H.G.; Ferreira, G.M.; Velozo, E.D.S.; Villarreal, C.F.; et al. The anti-inflammatory and immunomodulatory potential of braylin: Pharmacological properties and mechanisms by in silico, in vitro and in vivo approaches. PLoS ONE 2017, 12, e0179174. [Google Scholar] [CrossRef] [PubMed]
- Wittine, K.; Antolović, R.; Jelić, D.; Bracanović, S.; Cetina, M.; Andjelkovic, U.; Wittine, O.; Kraljević, P.S.; Vinter, A. Thienochromene derivatives inhibit pSTAT1 and pSTAT5 signaling induced by cytokines. Bioorg. Med. Chem. Lett. 2020, 30, 127415. [Google Scholar] [CrossRef] [PubMed]
- Gotō, H.; Ōsaw, E. An efficient algorithm for searching low-energy conformers of cyclic and acyclic molecules. J. Chem. Soc. Perkin Trans. 1993, 2, 187–198. [Google Scholar] [CrossRef]
- Jiang, L.; Ma, X.; Wang, Y.; Yang, J.P.; Huang, Y.; Liu, C.H.; Li, Y.J. New monoterpenoid glycosides from the fruits of Hypericum patulum thunb. Molecules 2024, 29, 3075. [Google Scholar] [CrossRef]
- Harakeh, S.; Niyazi, H.A.; Abdalal, S.A.; Mokhtar, J.A.; Almuhayawi, M.S.; Alkuwaity, K.K.; Abujamel, T.S.; Slama, P.; Haque, S. Integrated network pharmacology approach to evaluate bioactive phytochemicals of acalypha indica and their mechanistic actions to suppress target genes of tuberculosis. ACS Omega 2024, 9, 2204–2219. [Google Scholar] [CrossRef]
- Shi, P.X.; Chen, J.X.; Ge, W.Y.; Liu, Z.H.; Han, N.; Yin, J. Antichilblain components in eggplant based on network pharmacology and biological evaluation. J. Agric. Food Chem. 2023, 71, 11442–11453. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhong, S.X.; Tan, H.S.; Fu, Y.F.; Lai, J.Y.; Liu, L.J.; Weng, J.L.; Chen, H.W.; He, S.H. Study on the mechanism of action of Saposhnikovia divaricata and its key phytochemical on rheumatoid arthritis based on network pharmacology and bioinformatics. J. Ethnopharmacol. 2024, 322, 117586. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, F.; Nie, J. Anti-rheumatic effects of aconitum leucostomum worosch. on human fibroblast-like synoviocyte rheumatoid arthritis cells. Exp. Ther. Med. 2017, 14, 453. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
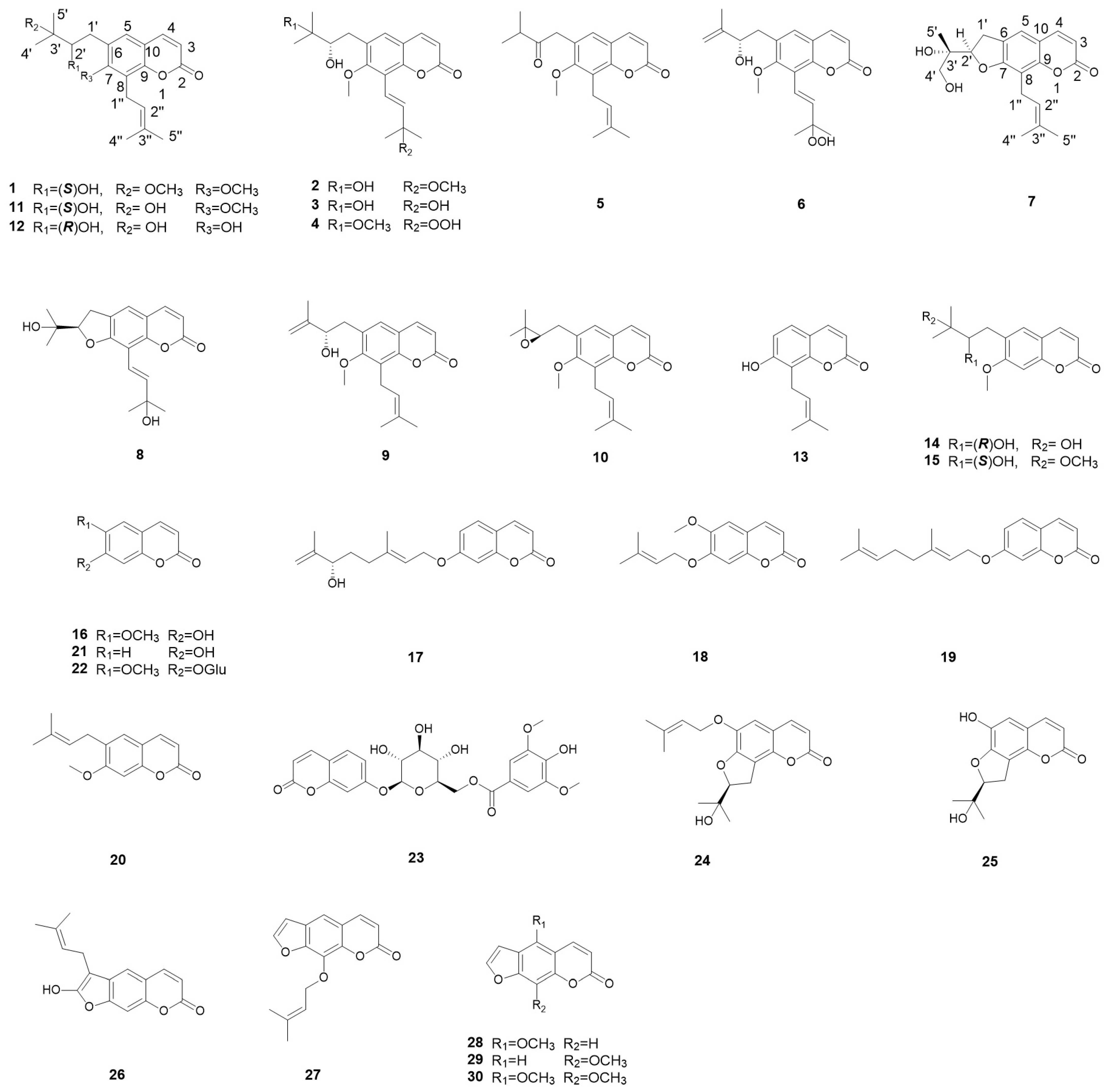
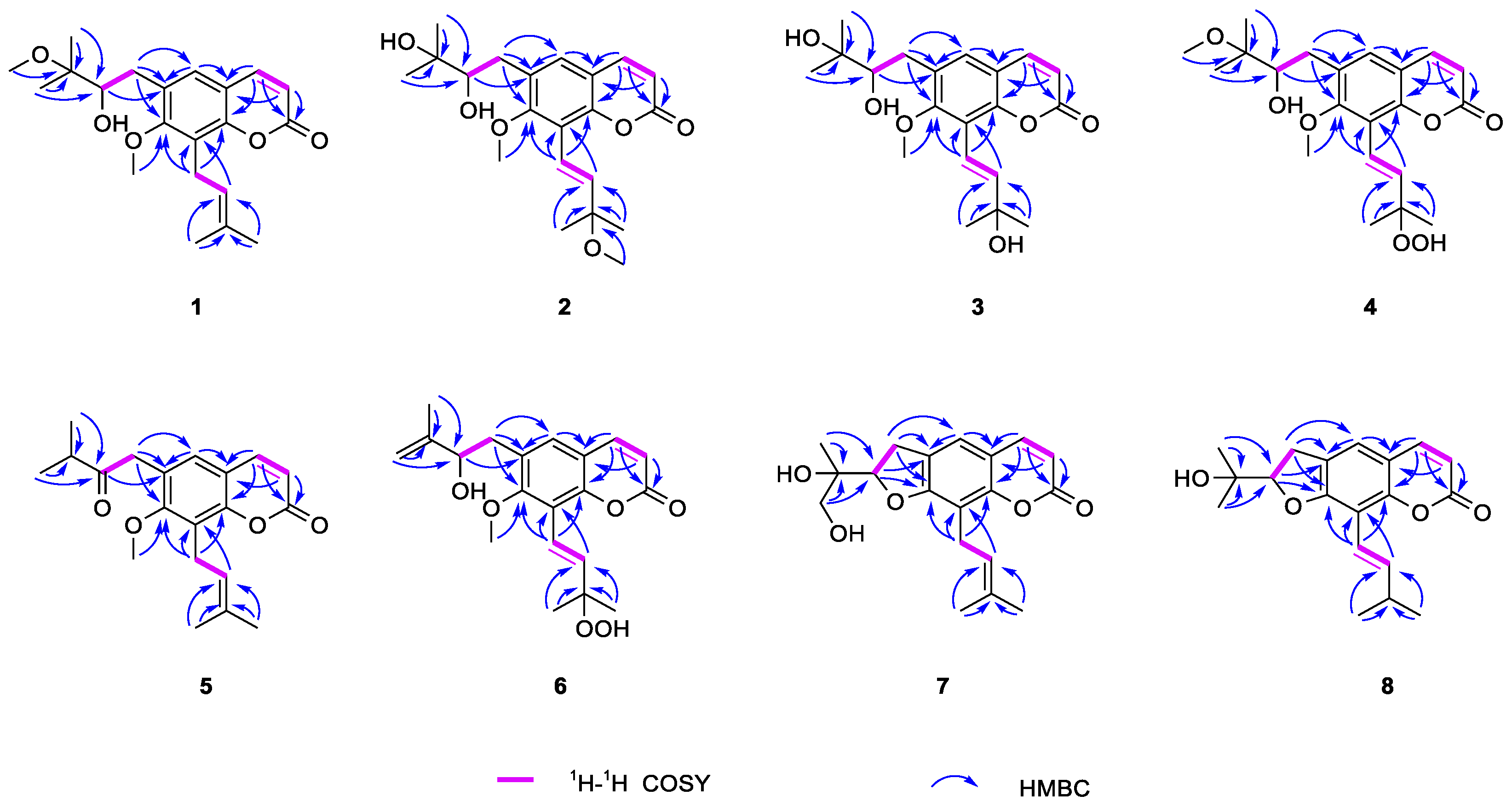

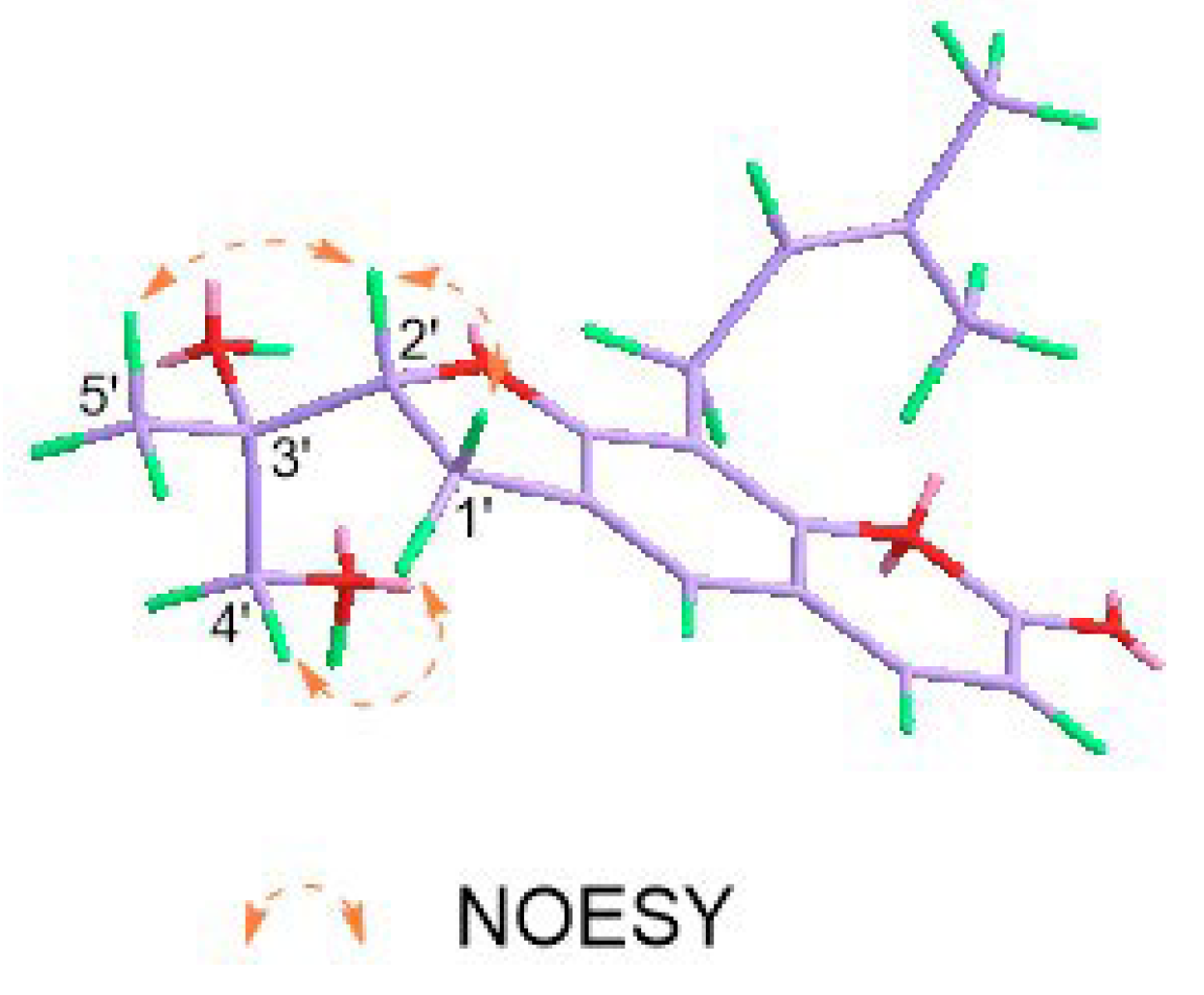




| Position | 1 b | 2 b | 2 a | 3 b | 4 a |
|---|---|---|---|---|---|
| 3 | 6.30 d (9.6) | 6.34 d (9.6) | 6.34 d (9.6) | 6.32 d (9.6) | 6.21 d (9.6) |
| 4 | 7.86 d (9.6) | 7.88 d (9.6) | 7.63 d (9.6) | 7.87 d (9.6) | 7.59 d (9.6) |
| 5 | 7.43 s | 7.47 s | 7.28 s | 7.44 s | 7.30 s |
| 1′α | 2.52 dd (14.4, 10.8) | 2.54 dd (14.4, 10.8) | 2.65 dd (14.4, 10.8) | 2.52 dd (14.4, 10.8) | 2.59 dd (14.4, 10.8) |
| 1′β | 3.09 dd (14.4, 2.4) | 3.12 dd (14.4, 2.4) | 2.95 dd (14.4, 1.8) | 3.12 dd (14.4, 1.8) | 2.96 dd (14.4, 2.4) |
| 2′ | 3.70 dd (10.8, 2.4) | 3.59 dd (10.8, 2.4) | 3.63 brd (10.8) | 3.58 dd (10.8, 1.8) | 3.83 dd (10.8, 2.4) |
| 3′ | |||||
| 4′ | 1.25 s | 1.28 s | 1.33 s | 1.28 s | 1.30 s |
| 5′ | 1.23 s | 1.27 s | 1.30 s | 1.27 s | 1.29 s |
| 1″α | 3.50 dd (14.4, 6.6) | 6.74 overlapped | 6.71 d (16.8) | 6.82 d (16.8) | 6.61 d (16.6) |
| 1″β | 3.56 dd (14.4, 6.6) | ||||
| 2″ | 5.22 t (6.6) | 6.74 overlapped | 6.77 d (16.8) | 6.92 d (16.8) | 6.86 d (16.6) |
| 4″ | 1.84 s | 1.43 s | 1.43 s | 1.43 s | 1.50 s |
| 5″ | 1.68 s | 1.43 s | 1.43 s | 1.44 s | 1.48 s |
| OCH3−7 | 3.82 s | 3.80 s | 3.78 s | 3.80 s | 3.76 s |
| OCH3−3′ | 3.30 s | 3.32 s | |||
| OCH3−3″ | 3.30 s | 3.28 s | |||
| OH−2′ | 2.61 brs | 2.56 brs | |||
| OH−3′ | 2.62 brs overlapped | 2.14 brs | |||
| OOH | 9.80 brs |
| Position | 1 b | 2 b | 3 b | 4 a | 5 a | 6 a | 7 b | 8 b |
|---|---|---|---|---|---|---|---|---|
| 2 | 163.2 | 162.8 | 163.0 | 161.5 | 160.9 | 161.5 | 164.0 | 163.6 |
| 3 | 114.9 | 115.1 | 114.9 | 114.0 | 115.0 | 115.2 | 111.6 | 111.9 |
| 4 | 146.0 | 146.1 | 146.2 | 144.4 | 143.4 | 144.4 | 146.8 | 146.8 |
| 5 | 129.5 | 130.5 | 130.1 | 129.0 | 127.8 | 129.1 | 122.4 | 123.0 |
| 6 | 132.2 | 132.3 | 132.2 | 130.2 | 125.1 | 129.3 | 132.3 | 127.0 |
| 7 | 161.4 | 161.3 | 161.3 | 159.7 | 159.6 | 159.7 | 163.2 | 162.6 |
| 8 | 124.2 | 119.6 | 119.9 | 118.2 | 123.6 | 118.4 | 112.2 | 109.4 |
| 9 | 153.0 | 152.4 | 152.4 | 150.8 | 152.6 | 150.9 | 154.2 | 153.3 |
| 10 | 116.8 | 116.8 | 116.7 | 115.2 | 115.5 | 115.2 | 114.2 | 114.1 |
| 1′ | 32.8 | 32.9 | 32.9 | 31.5 | 41.5 | 36.2 | 30.2 | 30.1 |
| 2′ | 78.6 | 79.4 | 79.4 | 76.2 | 211.7 | 74.9 | 88.8 | 92.5 |
| 3′ | 77.5 | 73.8 | 73.8 | 77.3 | 40.8 | 147.0 | 74.8 | 72.2 |
| 4′ | 20.5 | 25.9 | 25.9 | 20.3 | 18.4 | 111.0 | 67.6 | 25.8 |
| 5′ | 21.7 | 24.9 | 25.0 | 20.4 | 18.4 | 18.2 | 20.5 | 24.3 |
| 1″ | 23.8 | 119.3 | 116.0 | 118.2 | 23.1 | 117.9 | 23.2 | 115.2 |
| 2″ | 122.9 | 143.4 | 146.2 | 142.0 | 121.4 | 142.3 | 122.1 | 145.0 |
| 3″ | 133.5 | 77.2 | 71.9 | 82.6 | 132.9 | 82.6 | 133.8 | 71.9 |
| 4″ | 18.2 | 26.2 | 29.9 | 24.4 | 18.4 | 24.4 | 18.1 | 30.1 |
| 5″ | 25.8 | 26.1 | 29.9 | 24.5 | 25.7 | 24.5 | 25.9 | 30.0 |
| 7−OCH3 | 62.2 | 61.6 | 61.4 | 60.9 | 61.8 | 61.1 | ||
| 3′−OCH3 | 49.7 | 49.4 | ||||||
| 3″−OCH3 | 51.0 |
| Position | 5 a | 6 a | 7 b | 8 b |
|---|---|---|---|---|
| 3 | 6.35 d (9.6) | 6.21 d (9.6) | 6.15 d (9.6) | 6.24 d (9.6) |
| 4 | 7.63 d (9.6) | 7.59 d (9.6) | 7.80 d (9.6) | 7.85 d (9.6) |
| 5 | 7.15 s | 7.26 s | 7.47 s | 7.3 s |
| 1′α | 3.84 s | 2.95 dd (13.8, 3.6) | 3.35 dd (15.6, 9.0) | 3.28 ddd (15.0, 9.0, 1.2) |
| 1′β | 2.79 dd (13.8, 9.6) | 3.24 dd (15.6, 9.0) | 3.23 ddd (15.0, 9.0, 1.2) | |
| 2′ | 4.43 dd (9.6, 3.6) | 4.94 t (9.0) | 4.83 dd (9.0, 7.8) | |
| 3′ | 2.78 m | |||
| 4′ | 1.19 d (7.2) | 5.07 s | 3.55 d (11.4) | 1.36 s |
| 4.90 s | 3.74 d (10.8) | |||
| 5′ | 1.19 d (7.2) | 1.88 s | 1.2 s | 1.30 s |
| 1″α | 3.56 d (6.6) | 6.62 d (16.8) | 3.46 brd (7.2) | 7.00 d (16.8) |
| 1″β | ||||
| 2″ | 5.24 t (6.6) | 6.86 d (16.8) | 5.28 t (7.2) | 7.08 d (16.8) |
| 4″ | 1.84 s | 1.48 s | 1.83 s | 1.44 s |
| 5″ | 1.69 s | 1.51 s | 1.67 s | 1.44 s |
| OCH3−7 | 3.74 s | 3.77 s | ||
| OOH | 9.65 | |||
| OH−3′ | 4.59 brs |
| Compound | Target | Binding Energy (Kcal/mol) |
|---|---|---|
| 7 | JUN | −5.3 |
| 7 | SRC | −8.5 |
| 5 | SRC | −8.5 |
| 1 | EGFR | −6.1 |
| 5 | EGFR | −6.0 |
| 7 | EGFR | −6.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, C.; Li, X.; Chen, J.; Luo, L.; Yuan, C.; Yang, J.; Hao, X.; Gu, W. Discovery of Coumarins from Zanthoxylum dimorphophyllum var. spinifoliumas and Their Potential against Rheumatoid Arthritis. Molecules 2024, 29, 4395. https://doi.org/10.3390/molecules29184395
Du C, Li X, Chen J, Luo L, Yuan C, Yang J, Hao X, Gu W. Discovery of Coumarins from Zanthoxylum dimorphophyllum var. spinifoliumas and Their Potential against Rheumatoid Arthritis. Molecules. 2024; 29(18):4395. https://doi.org/10.3390/molecules29184395
Chicago/Turabian StyleDu, Caixia, Xingyu Li, Junlei Chen, Lili Luo, Chunmao Yuan, Jue Yang, Xiaojiang Hao, and Wei Gu. 2024. "Discovery of Coumarins from Zanthoxylum dimorphophyllum var. spinifoliumas and Their Potential against Rheumatoid Arthritis" Molecules 29, no. 18: 4395. https://doi.org/10.3390/molecules29184395
APA StyleDu, C., Li, X., Chen, J., Luo, L., Yuan, C., Yang, J., Hao, X., & Gu, W. (2024). Discovery of Coumarins from Zanthoxylum dimorphophyllum var. spinifoliumas and Their Potential against Rheumatoid Arthritis. Molecules, 29(18), 4395. https://doi.org/10.3390/molecules29184395







