Bioactive Compounds Produced by Endophytic Bacteria and Their Plant Hosts—An Insight into the World of Chosen Herbaceous Ruderal Plants in Central Europe
Abstract
:1. Introduction
2. Endophytes and Their Sources
3. Endophytic Plant Colonization Mechanisms
3.1. Physiological Mechanisms
3.2. Molecular Mechanisms
4. Interaction between Plants and Endophytic Microorganisms
5. Surface Sterilization of Plant Material
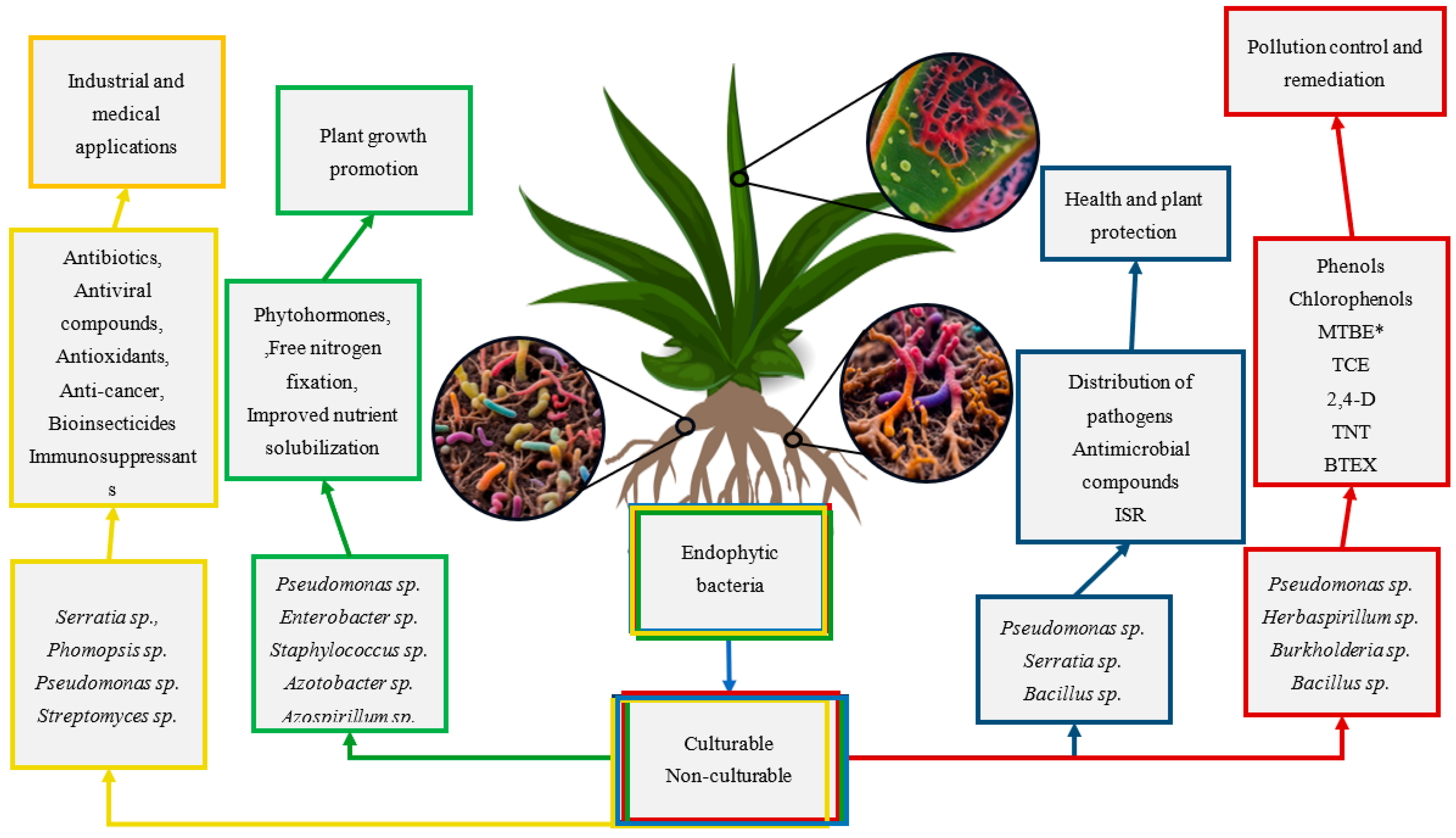
6. Bioactive Metabolites from Endophytic Bacteria
Genomics for Synthesis of Bioactive Compounds in Endophytic Bacteria
7. Ruderal Plants
7.1. Ruderal Herbaceous Plants
7.2. Biotechnological Potential of Endophytic Bacteria from Ruderal Herbaceous Plants
7.2.1. Chelidonium majus L.
7.2.2. Urtica dioica L.
7.2.3. Plantago lanceolata L.
7.2.4. Matricaria chamomilla L.
7.2.5. Equisetum arvense L.
7.2.6. Oenothera biennis L.
7.2.7. Silybum marianum L.
7.2.8. Mentha piperita L.
8. Future Perspectives
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, D. Endophyte: The Evolution of a Term, and Clarification of Its Use and Definition. Oikos 1995, 73, 274. [Google Scholar] [CrossRef]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef]
- Vandana, U.K.; Rajkumari, J.; Singha, L.P.; Satish, L.; Alavilli, H.; Sudheer, P.D.V.N.; Chauhan, S.; Ratnala, R.; Satturu, V.; Mazumder, P.B.; et al. The Endophytic Microbiome as a Hotspot of Synergistic Interactions, with Prospects of Plant Growth Promotion. Biology 2021, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Alvin, A.; Miller, K.I.; Neilan, B.A. Exploring the potential of endophytes from medicinal plants as sources of antimycobacterial compounds. Microbiol. Res. 2014, 169, 483–495. [Google Scholar] [CrossRef]
- Backman, P.A.; Sikora, R.A. Endophytes: An emerging tool for biological control. Biol. Control 2008, 46, 1–3. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, L.; Li, L.; Zheng, C.; Guo, L.; Li, W.; Sun, P.; Qin, L. Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiol. Res. 2010, 165, 437–449. [Google Scholar] [CrossRef]
- Thakur, D.; Sahani, K. In Vitro Antimicrobial Activity and Mic of the Extracellular Ethyl Acetate Crude Extract of Endophytic Fungi fusarium sp. Isolated From Tephrosia Purpurea Root. Int. J. Pharm. Pharm. Sci. 2019, 11, 48–53. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef]
- Marchut-Mikolajczyk, O.; Drozdzynski, P. Endophytic Microorganisms from Synanthropic Plants. In Bioremediation Science from Theory to Practice; CRC Press: Boca Raton, FL, USA, 2021; pp. 318–329. [Google Scholar]
- Pereira, S.I.A.; Monteiro, C.; Vega, A.L.; Castro, P.M.L. Endophytic culturable bacteria colonizing Lavandula dentata L. plants: Isolation, characterization and evaluation of their plant growth-promoting activities. Ecol. Eng. 2016, 87, 91–97. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, R.; Yadav, A.; Giri, D.D.; Singh, P.K.; Pandey, K.D. Isolation and characterization of bacterial endophytes of Curcuma longa L. 3 Biotech. 2016, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Nagda, V.; Gajbhiye, A.; Kumar, D. Isolation and Characterization of Endophytic fungi from Calotropis Procera for Their Antioxidant Activity. Asian, J. Pharm. Clin. Res. 2017, 10, 254–258. [Google Scholar] [CrossRef]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Surjit, S.D.; Rupa, G. Beneficial properties, colonization, establishment and molecular diversity of endophytic bacteria in legumes and non legumes. Afr. J. Microbiol. Res. 2014, 8, 1562–1572. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef]
- Elbeltagy, A.; Nishioka, K.; Sato, T.; Suzuki, H.; Ye, B.; Hamada TIsawa, T.; Mitsui, H.; Minamisawa, K. Endophytic Colonization and in Planta Nitrogen Fixation by a Herbaspirillum sp. Isolated from Wild Rice Species. Appl. Environ. Microbiol. 2001, 67, 5285–5293. [Google Scholar] [CrossRef]
- Afzal, M.; Khan, Q.M.; Sessitsch, A. Endophytic bacteria: Prospects and applications for the phytoremediation of organic pollutants. Chemosphere 2014, 117, 232–242. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil. Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Klama, J. Współśycie Endofitów Bakteryjnych z Roślinami (Artykuł Przeglądowy). Acta Sci. Pol. 2004, 3, 19–28. [Google Scholar]
- Malfanova, N.; Lugtenberg, B.J.J.; Berg, G. Bacterial Endophytes: Who and Where, and What Are They Doing There? In Molecular Microbial Ecology of the Rhizosphere; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Pisarksa, K.; Pietr, S. Bakterie endofityczne—Ich Pochodzenie i Interakcje z Roślinami. Post. Mikrobiol. 2014, 53, 141–151. [Google Scholar]
- Santoyo, G.; Urtis-Flores, C.A.; Loeza-Lara, P.D.; Orozco-Mosqueda, M.D.C.; Glick, B.R. Rhizosphere Colonization Determinants by Plant Growth-Promoting Rhizobacteria (PGPR). Biology 2021, 10, 475. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Oksinska, M.P.; Wright, S.A.I.; Pietr, S.J. Colonization of wheat seedlings (Triticum aestivum L.) by strains of Pseudomonas spp. with respect to their nutrient utilization profiles. Eur. J. Soil. Biol. 2011, 47, 364–373. [Google Scholar] [CrossRef]
- Yaron, S.; Römling, U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 2014, 7, 496. [Google Scholar] [CrossRef]
- James, E.K.; Olivares, F.L.; De Oliveira, A.L.M.; Reis, F.B.D.; Da Silva, L.G.; Reis, V.M. Further observations on the interaction between sugar cane and Gluconacetobacter diazotrophicus under laboratory and greenhouse conditions. J. Exp. Bot. 2001, 52, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Asaf, S.; Lee, I.J. What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Front. Plant Sci. 2018, 9, 24. [Google Scholar] [CrossRef]
- Oukala, N.; Pastor, V.; Aissat, K. Bacterial Endophytes: The Hidden Actor in Plant Immune Responses against Biotic Stress. Plants 2021, 10, 1012. [Google Scholar] [CrossRef]
- Frank, A.C.; Guzmán, J.P.S.; Shay, J.E. Transmission of Bacterial Endophytes. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.S.; Patra, J.K. Endophytes: A treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 2016, 7, 01538. [Google Scholar] [CrossRef]
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, S.; Shafiq, M.; Tariq, M.R.; Sami, A.; Nawaz-ul-Rehman, M.S.; Bhatti, M.H.T.; Haider, M.S.; Sadiq, S.; Abbas, M.T.; Hussain, M.; et al. Interaction between bacterial endophytes and host plants. Front. Plant Sci. 2023, 13, 1092105. [Google Scholar] [CrossRef] [PubMed]
- Bacilio-Jiménez, M.; Aguilar-Flores, S.; Ventura-Zapata, E.; Pérez-Campos, E.; Bouquelet, S.; Zenteno, E. Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 2003, 249, 271–277. [Google Scholar] [CrossRef]
- Bernardi, D.I.; Chagas, F.O.D.; Monteiro, A.F.; Santos, G.F.D.; de Souza Berlinck, R.G. Secondary Metabolites of Endophytic Actinomycetes: Isolation, Synthesis, Biosynthesis, and Biological Activities. Prog. Chem. Org. Nat. Prod. 2019, 108, 207–296. [Google Scholar] [CrossRef]
- Balachandar, D.; Sandhiya, G.S.; Sugitha, T.C.K.; Kumar, K. Flavonoids and growth hormones influence endophytic colonization and in planta nitrogen fixation by a diazotrophic Serratia sp. in rice. World J. Microbiol. Biotechnol. 2006, 22, 707–712. [Google Scholar] [CrossRef]
- Lee, K.-B.; De Backer, P.; Aono, T.; Liu, C.-T.; Suzuki, S.; Suzuki, T.; Kaneko, T.; Yamada, M.; Tabata, S.; Kupfer, D.M.; et al. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genom. 2008, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Pinski, A.; Betekhtin, A.; Hupert-Kocurek, K.; Mur, L.A.J.; Hasterok, R. Molecular Sciences Defining the Genetic Basis of Plant-Endophytic Bacteria Interactions. Int. J. Mol. Sci. 2019, 20, 1947. [Google Scholar] [CrossRef]
- de Weert, S.; Vermeiren, H.; Mulders, I.H.M.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J.J. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.A.; Medeot, D.B.; Liaudat, J.P.; Pistorio, M.; Jofré, E. In Azospirillum brasilense, mutations in flmA or flmB genes affect polar flagellum assembly, surface polysaccharides, and attachment to maize roots. Microbiol. Res. 2016, 190, 55–62. [Google Scholar] [CrossRef]
- Duque, E.; de la Torre, J.; Bernal, P.; Molina-Henares, M.A.; Alaminos, M.; Espinosa-Urgel, M.; Roca, A.; Fernández, M.; de Bentzmann, S.; Ramos, J.-L. Identification of reciprocal adhesion genes in pathogenic and non-pathogenic Pseudomonas. Environ. Microbiol. 2013, 15, 36–48. [Google Scholar] [CrossRef]
- Knights, H.E.; Jorrin, B.; Haskett, T.L.; Poole, P.S. Deciphering bacterial mechanisms of root colonization. Environ. Microbiol. Rep. 2021, 13, 428. [Google Scholar] [CrossRef] [PubMed]
- Balsanelli, E.; Tadra-Sfeir, M.Z.; Faoro, H.; Pankievicz, V.C.S.; de Baura, V.A.; Pedrosa, F.O.; de Souza, E.M.; Dixon, R.; Monteiro, R.A. Molecular adaptations of Herbaspirillum seropedicae during colonization of the maize rhizosphere. Environ. Microbiol. 2016, 18, 2343–2356. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, 0020396. [Google Scholar] [CrossRef] [PubMed]
- Fontana, D.C.; de Paula, S.; Torres, A.G.; de Souza, V.H.M.; Pascholati, S.F.; Schmidt, D.; Dourado Neto, D. Endophytic fungi: Biological control and induced resistance to phytopathogens and abiotic stresses. Pathogens 2021, 10, 570. [Google Scholar] [CrossRef]
- Bashir, I.; War, A.F.; Rafiq, I.; Reshi, Z.A.; Rashid, I.; Shouche, Y.S. Phyllosphere microbiome: Diversity and functions. Microbiol. Res. 2021, 254, 126888. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, C.; Liang, W.; Liu, S. Rhizosphere Microbiome: The Emerging Barrier in Plant-Pathogen Interactions. Front. Microbiol. 2021, 12, 772420. [Google Scholar] [CrossRef]
- Manghwar, H.; Zaman, W.; Kumari, M.; Swarupa, P.; Kesari, K.K.; Kumar, A. Microbial Inoculants as Plant Biostimulants: A Review on Risk Status. Life 2022, 13, 12. [Google Scholar] [CrossRef]
- Sahu, P.K.; Tilgam, J.; Mishra, S.; Hamid, S.; Gupta, A.; Verma, S.K.; Kharwar, R.N. Surface sterilization for isolation of endophytes: Ensuring what (not) to grow. J. Basic. Microbiol. 2022, 62, 647–668. [Google Scholar] [CrossRef]
- Yusof, H.; Saleh, N.N.; Sarmin, I.M. Optimization of the Isolation Method of Endophytic Actinomycetes from Psidium guajava (L.) and Ziziphus mauritiana/Hartini Yusof, Nurul Najwa Saleh and Nurul Izzah Mohd Sarmin. 2020. Available online: http://healthscopefsk.com/ (accessed on 11 April 2023).
- Schulz, B.; Wanke, U.; Draeger, S.; Aust, H.J. Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycol. Res. 1993, 97, 1447–1450. [Google Scholar] [CrossRef]
- Bhore, S.J.; Komathi, V.; Kandasamy, K.I. Diversity of endophytic bacteria in medicinally important Nepenthes species. J. Nat. Sci. Biol. Med. 2013, 4, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, L.T.; Gusmão, L.F.P.; Rodrigues, A. Diversity of endophytic fungi in Eucalyptus microcorys assessed by complementary isolation methods. Mycol. Prog. 2018, 17, 719–727. [Google Scholar] [CrossRef]
- Perez-Rosales, E.; Alcaraz-Meléndez, L.; Puente, M.E.; Vázquez-Juárez, R.; Quiroz-Guzmán, E.; Zenteno-Savín, T.; Morales-Bojórquez, E. Isolation and characterization of endophytic bacteria associated with roots of jojoba (Simmondsia chinensis (Link) Schneid). Curr. Sci. 2017, 112, 396–401. [Google Scholar]
- Compant, S.; Kaplan, H.; Sessitsch, A.; Nowak, J.; Barka, E.A.; Clément, C. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: From the rhizosphere to inflorescence tissues. FEMS Microbiol. Ecol. 2008, 63, 84–93. [Google Scholar] [CrossRef]
- Jahromi, R.; Mogharab, V.; Jahromi, H.; Avazpour, A. Synergistic effects of anionic surfactants on coronavirus (SARS-CoV-2) virucidal efficiency of sanitizing fluids to fight COVID-19. Food Chem. Toxicol. 2020, 145, 111702. [Google Scholar] [CrossRef]
- Xiang, H.; Zhang, T.; Pang, X.; Wei, Y.; Liu, H.; Zhang, Y.; Ma, B.; Yu, L. Isolation of endophytic fungi from Dioscorea zingiberensis C. H. Wright and application for diosgenin production by solid-state fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 5519–5532. [Google Scholar] [CrossRef]
- Jaber, L.R.; Enkerli, J. Effect of seed treatment duration on growth and colonization of Vicia faba by endophytic Beauveria bassiana and Metarhizium brunneum. Biol. Control 2016, 103, 187–195. [Google Scholar] [CrossRef]
- Rutala, W.A.; Weber, D.J. Disinfection, Sterilization, and Control of Hospital Waste. Mand. Douglas Bennett’s Princ. Pract. Infect. Dis. 2015, 2, 3294. [Google Scholar] [CrossRef]
- Munif, A.; Hallmann, J.; Sikora, R.A. Isolation of root endophytic bacteria from tomato and its biocontrol activity againts fungal diseases. Microbiol. Indones. 2012, 6, 2. [Google Scholar] [CrossRef]
- Misaghi, I.J. Endophytic Bacteria in Symptom-Free Cotton Plants. Phytopathology 1990, 80, 808. [Google Scholar] [CrossRef]
- Vega, F.E.; Pava-Ripoll, M.; Posada, F.; Buyer, J.S. Endophytic bacteria in Coffea arabica L. J. Basic. Microbiol. 2005, 45, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Mcdonnell, G.; Russell, A.D. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Thomas, P.; Singh, S.; Gupta, A. Taxonomic and functional diversity of cultivable endophytes with respect to the fitness of cultivars against Ralstonia solanacearum. J. Plant Dis. Prot. 2020, 127, 667–676. [Google Scholar] [CrossRef]
- Sahu, P.K.; Singh, S.; Gupta, A.; Singh, U.B.; Paul, S.; Paul, D.; Kuppusamy, P.; Singh, H.V.; Saxena, A.K. A simplified protocol for reversing phenotypic conversion of Ralstonia solanacearum during experimentation. Int. J. Environ. Res. Public Health 2020, 17, 4274. [Google Scholar] [CrossRef]
- Chen, J.; Akutse, K.S.; Saqib, H.S.A.; Wu, X.; Yang, F.; Xia, X.; Wang, L.; Goettel, M.S.; You, M.; Gurr, G.M. Fungal Endophyte Communities of Crucifer Crops Are Seasonally Dynamic and Structured by Plant Identity, Plant Tissue and Environmental Factors. Front. Microbiol. 2020, 11, 1519. [Google Scholar] [CrossRef]
- Teimoori-Boghsani, Y.; Ganjeali, A.; Cernava, T.; Müller, H.; Asili, J.; Berg, G. Endophytic Fungi of Native Salvia abrotanoides Plants Reveal High Taxonomic Diversity and Unique Profiles of Secondary Metabolites. Front. Microbiol. 2020, 10, 3013. [Google Scholar] [CrossRef]
- Agarwal, H.; Dowarah, B.; Baruah, P.M.; Bordoloi, K.S.; Krishnatreya, D.B.; Agarwala, N. Endophytes from Gnetum gnemon L. can protect seedlings against the infection of phytopathogenic bacterium Ralstonia solanacearum as well as promote plant growth in tomato. Microbiol. Res. 2020, 238, 126503. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, J.; Li, Y.; Wen, J.; Wang, R. Bacterial endophytes from Lycoris radiata promote the accumulation of Amaryllidaceae alkaloids. Microbiol. Res. 2020, 239, 126501. [Google Scholar] [CrossRef]
- Alibrandi, P.; Lo Monaco, N.; Calevo, J.; Voyron, S.; Puglia, A.M.; Cardinale, M.; Perotto, S. Plant growth promoting potential of bacterial endophytes from three terrestrial mediterranean orchid species. Plant Biosyst. 2021, 155, 1153–1164. [Google Scholar] [CrossRef]
- Ashitha, A.; Midhun, S.J.; Sunil, M.A.; Nithin, T.U.; Radhakrishnan, E.K.; Mathew, J. Bacterial endophytes from Artemisia nilagirica (Clarke) Pamp., with antibacterial efficacy against human pathogens. Microb. Pathog. 2019, 135, 103624. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhuang, X.; Yu, Z.; Wang, Z.; Wang, Y.; Guo, X.; Xiang, W.; Huang, S. Community Structures and Antifungal Activity of Root-Associated Endophytic Actinobacteria of Healthy and Diseased Soybean. Microorganisms 2019, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Zhou, Y.; Liu, H.; Yu, X. Endophytic actinomycetes from tea plants (Camellia sinensis): Isolation, abundance, antimicrobial, and plant-growth-promoting activities. Biomed. Res. Int. 2018, 2018, 1470305. [Google Scholar] [CrossRef] [PubMed]
- Moronta-Barrios, F.; Gionechetti, F.; Pallavicini, A.; Marys, E.; Venturi, V. Bacterial Microbiota of Rice Roots: 16S-Based Taxonomic Profiling of Endophytic and Rhizospheric Diversity, Endophytes Isolation and Simplified Endophytic Community. Microorganisms 2018, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Padda, K.P.; Puri, A.; Chanway, C.P. Isolation and identification of endophytic diazotrophs from lodgepole pine trees growing at unreclaimed gravel mining pits in central interior British Columbia, Canada. Can. J. For. Res. 2018, 48, 1601–1606. [Google Scholar] [CrossRef]
- Yaish, M.W.; Antony, I.; Glick, B.R. Isolation and characterization of endophytic plant growth-promoting bacteria from date palm tree (Phoenix dactylifera L.) and their potential role in salinity tolerance. Antonie Van Leeuwenhoek 2015, 107, 1519–1532. [Google Scholar] [CrossRef]
- Qin, S.; Li, J.; Chen, H.-H.; Zhao, G.-Z.; Zhu, W.-Y.; Jiang, C.-L.; Xu, L.-H.; Li, W.-J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna China. Appl. Environ. Microbiol. 2009, 75, 6176–6186. [Google Scholar] [CrossRef]
- Passari, A.K.; Mishra, V.K.; Saikia, R.; Gupta, V.K.; Singh, B.P. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 2015, 6, 273. [Google Scholar] [CrossRef]
- Nawed, A.; Chandra, R. Endophytic Bacteria: Optimizaton of Isolation Procedure from Various Medicinal Plants and Their Preliminary Characterization. Asian J. Pharm. Clin. Res. 2015, 8, 233–238. Available online: https://journals.innovareacademics.in/index.php/ajpcr/article/view/6586/2724 (accessed on 23 May 2023).
- Dukan, S.; Belkin, S.; Touati, D. Reactive oxygen species are partially involved in the bacteriocidal action of hypochlorous acid. Arch. Biochem. Biophys. 1999, 367, 311–316. [Google Scholar] [CrossRef]
- Le Dantec, C.; Duguet, J.P.; Montiel, A.; Dumoutier, N.; Dubrou, S.; Vincent, V. Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl. Environ. Microbiol. 2002, 68, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- ICSC 0979—MERCURIC CHLORIDE. Available online: https://www.inchem.org/documents/icsc/icsc/eics0979.htm (accessed on 6 August 2024).
- Gupta, R.; Kale, P.S.; Rathi, M.L.; Jadhav, N. Isolation, characterization and identification of endophytic bacteria by 16S rRNA partial sequencing technique from roots and leaves of Prosopis cineraria plant. Asian J. Plant Sci. Res. 2015, 5, 36–43. [Google Scholar]
- Gohain, A.; Gogoi, A.; Debnath, R.; Yadav, A.; Singh, B.P.; Gupta, V.K.; Sharma, R.; Saikia, R. Antimicrobial biosynthetic potential and genetic diversity of endophytic actinomycetes associated with medicinal plants. FEMS Microbiol. Lett. 2015, 362, 158. [Google Scholar] [CrossRef] [PubMed]
- Coombs, J.T.; Franco, C.M.M. Isolation and Identification of Actinobacteria from Surface-Sterilized Wheat Roots. Appl. Environ. Microbiol. 2003, 69, 5603. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.M.; Chandler, J.L.; Feldman, A.W. Growth promotion and inhibition by antibiotic-producing fluorescent pseudomonads on citrus roots. Plant Soil 1984, 77, 103–113. [Google Scholar] [CrossRef]
- Bell, C.R.; Dickie, G.A.; Harvey, W.L.G.; Chan, J.W.Y.F. Endophytic bacteria in grapevine. Can. J. Microbiol. 2011, 41, 46–53. [Google Scholar] [CrossRef]
- Scholander, P.F.; Hammel, H.T.; Bradstreet, E.D.; Hemmingsen, E.A. Sap Pressure in Vascular Plants: Negative hydrostatic pressure can be measured in plants. Science 1965, 148, 339–346. [Google Scholar] [CrossRef]
- Achari, G.A.; Ramesh, R. Diversity, Biocontrol, and Plant Growth Promoting Abilities of Xylem Residing Bacteria from Solanaceous Crops. Int. J. Microbiol. 2014, 2014, 296521. [Google Scholar] [CrossRef]
- Khare, E.; Mishra, J.; Arora, N.K. Multifaceted interactions between endophytes and plant: Developments and Prospects. Front. Microbiol. 2018, 9, 411604. [Google Scholar] [CrossRef]
- Poveda, J.; Díaz-González, S.; Díaz-Urbano, M.; Velasco, P.; Sacristán, S. Fungal endophytes of Brassicaceae: Molecular interactions and crop benefits. Front. Plant Sci. 2022, 13, 932288. [Google Scholar] [CrossRef]
- Gohain, A.; Gogoi, A.; Debnath, R.; Yadav, A.; Singh, B.P.; Gupta, V.K.; Sharma, R.; Saikia, R. Current Understanding of Endophytes: Their Relevance, Importance, and Industrial Potentials. IOSR J. Biotechnol. Biochem. 2017, 3, 43–59. [Google Scholar] [CrossRef]
- de Oliveira, N.C. Endophytic bacteria with potential for bioremediation of petroleum hydrocarbons and derivatives. Afr. J. Biotechnol. 2012, 11, ajb10.2623. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, C.C. Endophytes: A potential resource for biosynthesis, biotransformation, and biodegradation. Ann. Microbiol. 2011, 61, 207–215. [Google Scholar] [CrossRef]
- El-Shatoury, S.; El Kazzaz, W.; Elkraly, O. Antimicrobial Activities of Actinomycetes Inhabiting Achillea Fragrantissima (Family: Compositae). Egypt J. Nat. Toxins 2009, 6, 1–15. Available online: https://www.researchgate.net/publication/287492658 (accessed on 7 April 2024).
- Erjaee, Z.; Shekarforoush, S.S.; Hosseinzadeh, S. Identification of endophytic bacteria in medicinal plants and their antifungal activities against food spoilage fungi. J. Food Sci. Technol. 2019, 56, 5262–5270. [Google Scholar] [CrossRef]
- Rat, A.; Naranjo, H.D.; Krigas, N.; Grigoriadou, K.; Maloupa, E.; Varela Alonso, A.; Schneider, C.; Papageorgiou, V.P.; Assimopoulou, A.N.; Tsafantakis, N.; et al. Endophytic Bacteria From the Roots of the Medicinal Plant Alkanna tinctoria Tausch (Boraginaceae): Exploration of Plant Growth Promoting Properties and Potential Role in the Production of Plant Secondary Metabolites. Front. Microbiol. 2021, 12, 633488. [Google Scholar] [CrossRef]
- Igarashi, Y. Screening of Novel Bioactive Compounds from Plant-Associated Actinomycetes. Actinomycetologica 2004, 18, 63–66. [Google Scholar] [CrossRef]
- Sasaki, T.; Igarashi, Y.; Ogawa, M.; Furumai, T. Identification of 6-Prenylindole as an Antifungal Metabolite of Streptomyces sp. TP-A0595 and Synthesis and Bioactivity of 6-Substituted Indoles. J. Antibiot. 2002, 55, 1009–1012. [Google Scholar] [CrossRef]
- Liu, J.-Q.; Chen, S.-M.; Zhang, C.-M.; Xu, M.-J.; Xing, K.; Li, C.-G.; Li, K.; Zhang, Y.-Q.; Qin, S. Abundant and diverse endophytic bacteria associated with medicinal plant Arctium lappa L. and their potential for host plant growth promoting. Antonie Van Leeuwenhoek Int. J. General Mol. Microbiol. 2022, 115, 1405–1420. [Google Scholar] [CrossRef]
- Shurigin, V.; Egamberdieva, D.; Samadiy, S.; Mardonova, G.; Davranov, K. Endophytes from medicinal plants as biocontrol agents against fusarium caused diseases. Mikrobiolohichnyi Zhurnal 2020, 82, 41–52. [Google Scholar] [CrossRef]
- Wardecki, T.; Brötz, E.; De Ford, C.; von Loewenich, F.D.; Rebets, Y.; Tokovenko, B.; Luzhetskyy, A.; Merfort, I. Endophytic Streptomyces in the traditional medicinal plant Arnica montana L.: Secondary metabolites and biological activity. Antonie Van Leeuwenhoek Int. J. General Mol. Microbiol. 2015, 108, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Bekiesch, P.; Oberhofer, M.; Sykora, C.; Urban, E.; Zotchev, S.B. Piperazic acid containing peptides produced by an endophytic Streptomyces sp. isolated from the medicinal plant Atropa belladonna. Nat. Prod. Res. 2021, 35, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Krimi, Z.; Alim, D.; Djellout, H.; Tafifet, L.; Mohamed-Mahmoud, F.; Raio, A. Bacterial endophytes of weeds are effective biocontrol agents of Agrobacterium spp., Pectobacterium spp., and promote growth of tomato plants. Phytopathol. Mediterr. 2016, 55, 184–196. [Google Scholar] [CrossRef]
- Köberl, M.; Ramadan, E.M.; Adam, M.; Cardinale, M.; Hallmann, J.; Heuer, H.; Smalla, K.; Berg, G. Bacillus and streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in egypt. FEMS Microbiol. Lett. 2013, 342, 168–178. [Google Scholar] [CrossRef]
- Shurigin, V.; Alimov, J.; Davranov, K.; Gulyamova, T.; Egamberdieva, D. The diversity of bacterial endophytes from Iris pseudacorus L. and their plant beneficial traits. Curr. Res. Microb. Sci. 2022, 3, 100133. [Google Scholar] [CrossRef]
- Ait Kaki, A.; Kacem Chaouche, N.; Dehimat, L.; Milet, A.; Youcef-Ali, M.; Ongena, M.; Thonart, P. Biocontrol and Plant Growth Promotion Characterization of Bacillus Species Isolated from Calendula officinalis Rhizosphere. Indian. J. Microbiol. 2013, 53, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Marchut-Mikolajczyk, O.; Drozdzyński, P.; Pietrzyk, D.; Antczak, T. Biosurfactant production and hydrocarbon degradation activity of endophytic bacteria isolated from Chelidonium majus L. 06 Biological Sciences 0605 Microbiology 06 Biological Sciences 0607 Plant Biology 09 Engineering 0907 Environmental Engineering. Microb Cell Fact 2018, 17, 171. [Google Scholar] [CrossRef]
- Goryluk-Salmonowicz, A.; Orzeszko-Rywka, A.; Piorek, M.; Rekosz-Burlaga, H.; Otlowska, A.; Gozdowski, D.; Blaszczyk, M. Plant growth promoting bacterial endophytes isolated from Polish herbal plants. Acta Sci. Pol. Hortorum Cultus 2018, 17, 101–110. [Google Scholar] [CrossRef]
- Goryluk, A.; Rekosz-Burlaga, H.; Błaszczyk, M. Isolation and Characterization of Bacterial Endophytes of Chelidonium majus L. Pol. J. Microbiol. 2009, 58, 355–361. [Google Scholar]
- Presta, L.; Bosi, E.; Fondi, M.; Maida, I.; Perrin, E.; Miceli, E.; Maggini, V.; Bogani, P.; Firenzuoli, F.; Di Pilato, V.; et al. Phenotypic and genomic characterization of the antimicrobial producer Rheinheimera sp. EpRS3 isolated from the medicinal plant Echinacea purpurea: Insights into its biotechnological relevance. Res. Microbiol. 2017, 168, 293–305. [Google Scholar] [CrossRef]
- Goryluk-Salmonowicz, A.; Piórek, M.; Rekosz-Burlaga, H.; Studnicki, M.; Błaszczyk, M. Endophytic Detection in Selected European Herbal Plants. Pol. J. Microbiol. 2016, 65, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Patra, J.K.; Baek, K.H. Antibacterial Properties of Endophytic Bacteria Isolated from a Fern Species Equisetum arvense L. Against Foodborne Pathogenic Bacteria Staphylococcus aureus and Escherichia coli O157:H7. Foodborne Pathog. Dis. 2017, 14, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, M.; Gałązka, A.; Tyśkiewicz, R.; Jaroszuk-ściseł, J. Endophytic bacteria potentially promote plant growth by synthesizing different metabolites and their phenotypic/physiological profiles in the biolog gen iii microplateTM test. Int. J. Mol. Sci. 2019, 20, 5283. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Behrendt, U.; Ahmad, P.; Berg, G. Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front. Microbiol. 2017, 8, 00199. [Google Scholar] [CrossRef]
- Pawlik, M.; Cania, B.; Thijs, S.; Vangronsveld, J.; Piotrowska-Seget, Z. Hydrocarbon degradation potential and plant growth-promoting activity of culturable endophytic bacteria of Lotus corniculatus and Oenothera biennis from a long-term polluted site. Environ. Sci. Pollut. Res. 2017, 24, 19640–19652. [Google Scholar] [CrossRef]
- Alaylar, B. Isolation and Characterization of Culturable Endophytic Plant Growth Promoting Bacillus Species from Mentha longifolia L. Turk. J. Agric. For. 2021, 46, 73–82. [Google Scholar] [CrossRef]
- Ting, A.S.Y.; Akinsanya, M.A.; Goh, J.K.; Lim, S.P. Antimicrobial and Enzymatic Activities of Endophytic Bacteria Isolated from Mentha spicata (MINT). Malays. J. Microbiol. 2015, 11, 102–108. Available online: https://www.researchgate.net/publication/282651721 (accessed on 7 April 2024).
- Ngashangva, N.; Mukherjee, P.; Sharma, K.C.; Kalita, M.C.; Indira, S. Analysis of Antimicrobial Peptide Metabolome of Bacterial Endophyte Isolated From Traditionally Used Medicinal Plant Millettia pachycarpa Benth. Front. Microbiol. 2021, 12, 65689. [Google Scholar] [CrossRef] [PubMed]
- Chlebek, D.; Pinski, A.; Żur, J.; Michalska, J.; Hupert-Kocurek, K. Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens. Int. J. Mol. Sci. 2020, 21, 8740. [Google Scholar] [CrossRef]
- Ben Mefteh, F.; Chenari Bouket, A.; Daoud, A.; Luptakova, L.; Alenezi, F.; Gharsallah, N.; Belbahri, L. Metagenomic Insights and Genomic Analysis of Phosphogypsum and Its Associated Plant Endophytic Microbiomes Reveals Valuable Actors for Waste Bioremediation. Microorganisms 2019, 7, 382. [Google Scholar] [CrossRef]
- Albarano, L.; Esposito, R.; Ruocco, N.; Costantini, M. Genome mining as new challenge in natural products discovery. Mar. Drugs 2020, 18, 199. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Fischbach, M.A.; Gilbert, J.A.; Greenberg, E.P.; Loper, J.E.; Metcalf, W.W.; O’Connor, S.E.; Piel, J.; Raaijmakers, J.M.; Stachelhaus, T.; et al. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39 (Suppl. S2), gkr466. [Google Scholar] [CrossRef] [PubMed]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.-C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef]
- Castillo, U.F.; Strobel, G.A.; Mullenberg, K.; Condron, M.M.; Teplow, D.B.; Folgiano, V.; Gallo, M.; Ferracane, R.; Mannina, L.; Viel, S.; et al. Munumbicins E-4 and E-5: Novel broad-spectrum antibiotics from Streptomyces NRRL 3052. FEMS Microbiol. Lett. 2006, 255, 296–300. [Google Scholar] [CrossRef]
- Li, J.; Zhao, G.-Z.; Huang, H.-Y.; Qin, S.; Zhu, W.-Y.; Zhao, L.-X.; Xu, L.-H.; Zhang, S.; Li, W.-J. Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Antonie Van Leeuwenhoek 2012, 101, 515–527. [Google Scholar] [CrossRef]
- Taria, S.; Arora, A.; Alam, B.; Kumar, S.; Yadav, A.; Kumar, S.; Kumar, M.; Anuragi, H.; Kumar, R.; Meena, S.; et al. Introduction to Plant Nitrogen Metabolism. In Advances in Plant Nitrogen Metabolism; CRC Press: Boca Raton, FL, USA, 2022; pp. 1–18. [Google Scholar] [CrossRef]
- Maela, M.P.; van der Walt, H.; Serepa-Dlamini, M.H. The Antibacterial, Antitumor Activities, and Bioactive Constituents’ Identification of Alectra sessiliflora Bacterial Endophytes. Front. Microbiol. 2022, 13, 870821. [Google Scholar] [CrossRef]
- Narayanan, Z.; Glick, B.R. Secondary Metabolites Produced by Plant Growth-Promoting Bacterial Endophytes. Microorganisms 2022, 10, 2008. [Google Scholar] [CrossRef]
- Ancheeva, E.; Daletos, G.; Proksch, P. Bioactive Secondary Metabolites from Endophytic Fungi. Curr. Med. Chem. 2019, 27, 1836–1854. [Google Scholar] [CrossRef]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Chekan, J.R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021, 38, 2100–2129. [Google Scholar] [CrossRef]
- Kaul, S.; Gupta, S.; Ahmed, M.; Dhar, M.K. Endophytic fungi from medicinal plants: A treasure hunt for bioactive metabolites. Phytochem. Rev. 2012, 11, 487–505. [Google Scholar] [CrossRef]
- Rutkowska, N.; Drożdżyński, P.; Ryngajłło, M.; Marchut-Mikołajczyk, O. Plants as the Extended Phenotype of Endophytes—The Actual Source of Bioactive Compounds. Int. J. Mol. Sci. 2023, 24, 96. [Google Scholar] [CrossRef] [PubMed]
- Strobel, G.A. Endophytes as Sources of Bioactive Products. 2003. Available online: www.elsevier.com/locate/micinf (accessed on 7 April 2024).
- Ansari, A.A.; Gill, S.S.; Gill, R.; Lanza, G.R.; Newman, L. Phytoremediation: Management of Environmental Contaminants; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; Volume 6. [Google Scholar] [CrossRef]
- Kusari, S.; Spiteller, M. Are we ready for industrial production of bioactive plant secondary metabolites utilizing endophytes? Nat. Prod. Rep. 2011, 28, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Pratyusha, S. Phenolic Compounds in the Plant Development and Defense: An Overview. Available online: www.intechopen.com (accessed on 3 March 2024).
- Rosconi, F.; Davyt, D.; Martínez, V.; Martínez, M.; Abin-Carriquiry, J.A.; Zane, H.; Butler, A.; de Souza, E.M.; Fabiano, E. Identification and structural characterization of serobactins, a suite of lipopeptide siderophores produced by the grass endophyte Herbaspirillum seropedicae. Environ. Microbiol. 2013, 15, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, P. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 2010, 86, 1637–1645. [Google Scholar] [CrossRef]
- Misra, S.; Semwal, P.; Pandey, D.D.; Mishra, S.K.; Chauhan, P.S. Siderophore-Producing Spinacia Oleracea Bacterial Endophytes Enhance Nutrient Status and Vegetative Growth Under Iron-Deficit Conditions. J. Plant Growth Regul. 2024, 43, 1317–1330. [Google Scholar] [CrossRef]
- Maheshwari, R.; Bhutani, N.; Suneja, P. Screening and characterization of siderophore producing endophytic bacteria from Cicer arietinum and Pisum sativum plants. J. Appl. Biol. Biotechnol. 2019, 7, 7–14. [Google Scholar] [CrossRef]
- Marchut-Mikołajczyk, O.; Chlebicz, M.; Kawecka, M.; Michalak, A.; Prucnal, F.; Nielipinski, M.; Filipek, J.; Jankowska, M.; Perek, Z.; Drożdżyński, P.; et al. Endophytic bacteria isolated from Urtica dioica L.-preliminary screening for enzyme and polyphenols production. Microb. Cell Fact. 2023, 22, 169. [Google Scholar] [CrossRef]
- Zahoor, M.; Irshad, M.; Rahman, H.; Qasim, M.; Afridi, S.G.; Qadir, M.; Hussain, A. Alleviation of heavy metal toxicity and phytostimulation of Brassica campestris L. by endophytic Mucor sp. MHR-7. Ecotoxicol. Environ. Saf. 2017, 142, 139–149. [Google Scholar] [CrossRef]
- Cornu, J.Y.; Huguenot, D.; Jézéquel, K.; Lollier, M.; Lebeau, T. Bioremediation of copper-contaminated soils by bacteria. World J. Microbiol. Biotechnol. 2017, 33, 26. [Google Scholar] [CrossRef]
- Do, H.; Che, C.; Zhao, Z.; Wang, Y.; Li, M.; Zhang, X.; Zhao, X. Extracellular polymeric substance from Rahnella sp. LRP3 converts available Cu into Cu5(PO4)2(OH)4 in soil through biomineralization process. Environ. Pollut. 2020, 260, 114051. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Do, H.; Zhou, Y.; Li, Z.; Zhang, X.; Zhao, S.; Li, M.; Wu, D. Rahnella sp. LRP3 induces phosphate precipitation of Cu (II) and its role in copper-contaminated soil remediation. J. Hazard. Mater. 2019, 368, 133–140. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Ye, Z.; Yang, D.; Yan, J.; Xiao, L.; Zhong, T.; Yuan, M.; Cai, X.; Fang, Z.; Jing, Y. Characterization of endophytic Rahnella sp. JN6 from Polygonum pubescens and its potential in promoting growth and Cd, Pb, Zn uptake by Brassica napus. Chemosphere 2013, 90, 1960–1965. [Google Scholar] [CrossRef]
- Jing, Y.X.; Yan, J.L.; He, H.D.; Yang, D.J.; Xiao, L.; Zhong, T.; Yuan, M.; Cai, X.; Li, S.B. Characterization of Bacteria in the Rhizosphere Soils of Polygonum Pubescens and Their Potential in Promoting Growth and Cd, Pb, Zn Uptake by Brassica napus. Int. J. Phytoremediation 2014, 16, 321–333. [Google Scholar] [CrossRef]
- Tamariz-Angeles, C.; Huamán, G.D.; Palacios-Robles, E.; Olivera-Gonzales, P.; Castañeda-Barreto, A. Characterization of siderophore-producing microorganisms associated to plants from high-Andean heavy metal polluted soil from Callejón de Huaylas (Ancash, Perú). Microbiol. Res. 2021, 250, 126811. [Google Scholar] [CrossRef] [PubMed]
- Ulloa-Muñoz, R.; Olivera-Gonzales, P.; Castañeda-Barreto, A.; Villena, G.K.; Tamariz-Angeles, C. Diversity of endophytic plant-growth microorganisms from Gentianella weberbaueri and Valeriana pycnantha, highland Peruvian medicinal plants. Microbiol. Res. 2020, 233, 126413. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Chen, B.; Luo, S.; Wu, Y.; Ye, J.; Wang, Q.; Xu, X.; Pan, F.; Khan, K.Y.; Feng, Y.; Yang, X.; et al. The effects of the endophytic bacterium Pseudomonas fluorescens Sasm05 and IAA on the plant growth and cadmium uptake of Sedum alfredii hance. Front. Microbiol. 2017, 8, 2538. [Google Scholar] [CrossRef] [PubMed]
- Tavarideh, F.; Pourahmad, F.; Nemati, M. Diversity and antibacterial activity of endophytic bacteria associated with medicinal plant, Scrophularia striata. Vet. Res. Forum 2022, 13, 409–415. [Google Scholar] [CrossRef]
- Hammerschmidt, R. Induced disease resistance: How do induced plants stop pathogens? Physiol. Mol. Plant Pathol. 1999, 55, 77–84. [Google Scholar] [CrossRef]
- Walters, D.; Walsh, D.; Newton, A.; Lyon, G. Induced resistance for plant disease control: Maximizing the efficacy of resistance elicitors. Phytopathology 2005, 95, 1368–1373. [Google Scholar] [CrossRef] [PubMed]
- Dudeja, S.S.; Suneja-Madan, P.; Paul, M.; Maheswari, R.; Kothe, E. Bacterial endophytes: Molecular interactions with their hosts. J. Basic. Microbiol. 2021, 61, 475–505. [Google Scholar] [CrossRef] [PubMed]
- Kenshole, E.; Herisse, M.; Michael, M.; Pidot, S.J. Natural product discovery through microbial genome mining. Curr. Opin. Chem. Biol. 2021, 60, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Cheffi, M.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A.; Belbahri, L. Olea europaea l. Root endophyte bacillus velezensis oee1 counteracts oomycete and fungal harmful pathogens and harbours a large repertoire of secreted and volatile metabolites and beneficial functional genes. Microorganisms 2019, 7, 314. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wen, W.; Qin, M.; He, Y.; Xu, D.; Li, L. Biosynthetic Mechanisms of Secondary Metabolites Promoted by the Interaction Between Endophytes and Plant Hosts. Front. Media 2022, 11, 928967. [Google Scholar] [CrossRef]
- Kaul, S.; Sharma, T.; Dhar, M.K. ‘Omics’ tools for better understanding the plant–endophyte interactions. Front. Plant Sci. 2016, 7, 955. [Google Scholar] [CrossRef]
- Guo, D.-J.; Li, D.-P.; Singh, R.K.; Singh, P.; Sharma, A.; Verma, K.K.; Qin, Y.; Khan, Q.; Lu, Z.; Malviya, M.K.; et al. Differential Protein Expression Analysis of Two Sugarcane Varieties in Response to Diazotrophic Plant Growth-Promoting Endophyte Enterobacter roggenkampii ED5. Front. Plant Sci. 2021, 12, 727741. [Google Scholar] [CrossRef]
- Zgadzaj, R.; James, E.K.; Kelly, S.; Kawaharada, Y.; de Jonge, N.; Jensen, D.B.; Madsen, L.H.; Radutoiu, S. A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria. PLoS Genet. 2015, 11, e1005280. [Google Scholar] [CrossRef]
- Monteiro, R.A.; Balsanelli, E.; Tuleski, T.; Faoro, H.; Cruz, L.M.; Wassem, R.; de Baura, V.A.; Tadra-Sfeir, M.Z.; Weiss, V.; DaRocha, W.D.; et al. Genomic comparison of the endophyte Herbaspirillum seropedicae SmR1 and the phytopathogen Herbaspirillum rubrisubalbicans M1 by suppressive subtractive hybridization and partial genome sequencing. FEMS Microbiol. Ecol. 2012, 80, 441–451. [Google Scholar] [CrossRef]
- Tian, D.; Song, X.; Li, C.; Zhou, W.; Qin, L.; Wei, L.; Di, W.; Huang, S.; Li, B.; Huang, Q.; et al. Antifungal mechanism of Bacillus amyloliquefaciens strain GKT04 against Fusarium wilt revealed using genomic and transcriptomic analyses. Microbiologyopen 2021, 10, e1192. [Google Scholar] [CrossRef]
- Luo, Y.; Cobb, R.E.; Zhao, H. Recent advances in natural product discovery. Curr. Opin. Biotechnol. 2014, 30, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Segers, K.; Declerck, S.; Mangelings, D.; Heyden, Y.V.; Van Eeckhaut, A. Analytical techniques for metabolomic studies: A review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef] [PubMed]
- Armin, R.; Zühlke, S.; Mahnkopp-Dirks, F.; Winkelmann, T.; Kusari, S. Evaluation of Apple Root-Associated Endophytic Streptomyces pulveraceus Strain ES16 by an OSMAC-Assisted Metabolomics Approach. Front. Sustain. Food Syst. 2021, 5, 643225. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Media 2019, 10, 00294. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, B.O.; Van Lanen, S.G.; Baltz, R.H. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 2014, 41, 175–184. [Google Scholar] [CrossRef]
- Kudo, F.; Miyanaga, A.; Eguchi, T. Structural basis of the nonribosomal codes for nonproteinogenic amino acid selective adenylation enzymes in the biosynthesis of natural products. J. Ind. Microbiol. Biotechnol. 2019, 46, 515–536. [Google Scholar] [CrossRef]
- Iacovelli, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Nonribosomal peptide synthetases and their biotechnological potential in Penicillium rubens. J. Ind. Microbiol. Biotechnol. 2021, 48, 45. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef]
- Le Govic, Y.; Papon, N.; Le Gal, S.; Bouchara, J.P.; Vandeputte, P. Non-ribosomal Peptide Synthetase Gene Clusters in the Human Pathogenic Fungus Scedosporium apiospermum. Front. Microbiol. 2019, 10, 476594. [Google Scholar] [CrossRef]
- Gulick, A.M. Nonribosomal peptide synthetase biosynthetic clusters of ESKAPE pathogens. Nat. Prod. Rep. 2017, 34, 981–1009. [Google Scholar] [CrossRef]
- Chu, H.Y.; Wegel, E.; Osbourn, A. From hormones to secondary metabolism: The emergence of metabolic gene clusters in plants. Plant J. 2011, 66, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete Genome Sequence of the Model Actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L.; Ravel, J. Coelichelin, a New Peptide Siderophore Encoded by the Streptomyces coelicolor Genome: Structure Prediction from the Sequence Of Its Non-Ribosomal Peptide Synthetase. FEMS Microbiol. Lett. 2000, 187, 111–114. Available online: www.fems-microbiology.org (accessed on 7 April 2024). [CrossRef] [PubMed]
- Ikeda, H.; Shin-Ya, K.; Omura, S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 2014, 41, 233–250. [Google Scholar] [CrossRef]
- Malpartida, F.; Hopwood, D.A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature 1984, 309, 462–464. [Google Scholar] [CrossRef]
- Ali, M.A.; Lou, Y.; Hafeez, R.; Li, X.; Hossain, A.; Xie, T.; Lin, L.; Li, B.; Yin, Y.; Yan, J.; et al. Functional Analysis and Genome Mining Reveal High Potential of Biocontrol and Plant Growth Promotion in Nodule-Inhabiting Bacteria Within Paenibacillus polymyxa Complex. Front. Microbiol. 2021, 11, 618601. [Google Scholar] [CrossRef] [PubMed]
- Kaewkla, O.; Franco, C.M.M. Genome mining and description of Streptomyces albidus sp. nov., an endophytic actinobacterium with antibacterial potential. Antonie Van Leeuwenhoek Int. J. General Mol. Microbiol. 2021, 114, 539–551. [Google Scholar] [CrossRef]
- Tsalgatidou, P.C.; Thomloudi, E.-E.; Baira, E.; Papadimitriou, K.; Skagia, A.; Venieraki, A.; Katinakis, P. Integrated Genomic and Metabolomic Analysis Illuminates Key Secreted Metabolites Produced by the Novel Endophyte Bacillus halotolerans Cal.l.30 Involved in Diverse Biological Control Activities. Microorganisms 2022, 10, 399. [Google Scholar] [CrossRef]
- Jasim, B.; Sreelakshmi, K.S.; Mathew, J.; Radhakrishnan, E.K. Surfactin, Iturin, and Fengycin Biosynthesis by Endophytic Bacillus sp. from Bacopa monnieri. Microb. Ecol. 2016, 72, 106–119. [Google Scholar] [CrossRef]
- Li, J.; Qu, X.; He, X.; Duan, L.; Wu, G.; Bi, D.; Deng, Z.; Liu, W.; Ou, H.-Y. ThioFinder: A web-based tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS ONE 2012, 7, e45878. [Google Scholar] [CrossRef]
- Scheffler, R.J.; Colmer, S.; Tynan, H.; Demain, A.L.; Gullo, V.P. Antimicrobials, drug discovery, and genome mining. Appl. Microbiol. Biotechnol. 2013, 97, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.S.; Katsuyama, Y.; Bai, L.; Deng, Z.; Ohnishi, Y.; Kim, E.S. Genome engineering for microbial natural product discovery. Curr. Opin. Microbiol. 2018, 45, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; Méndez, C.; Salas, J.A. Strategies for the design and discovery of novel antibiotics using genetic engineering and genome mining. In Antimicrobial Compounds: Current Strategies and New Alternatives; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–25. [Google Scholar] [CrossRef]
- Nowak, A.; Nowak, S.; Nobis, M.; Nobis, A. A report on the conservation status of segetal weeds in Tajikistan. Weed Res. 2014, 54, 635–648. [Google Scholar] [CrossRef]
- Siciński, J.T. Flora i roślinność (szata roślinna) synantropijna wsi oraz małych miasteczek. Zesz. Wiej. 2012, 17, 256–271. [Google Scholar]
- Yalçinalp, E.; Meral, A. Ruderal Plants in Urban and Sub-Urban Walls and Roofs. J. Agric. Fac. Ege Univ. 2019, 56, 205–212. [Google Scholar] [CrossRef]
- Fanfarillo, E. Segetal Plant Biodiversity in Italy: Floristic and Coenological Analyses at Different Spatial and Time Scales. Available online: https://www.researchgate.net/publication/339540030 (accessed on 14 December 2023).
- Jackowiak, B. Man-Made Changes in the Flora and Vegetation of Poland: Current Review. Diversity 2023, 15, 618. [Google Scholar] [CrossRef]
- Kelcey, J.G.; Müller, N. Plants and Habitats of European Cities; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–6851. [Google Scholar]
- Hosseini, Z.; Zangari, G.; Carboni, M.; Caneva, G. Substrate preferences of ruderal plants in colonizing stone monuments of the pasargadae world heritage site, Iran. Sustainability 2021, 13, 9381. [Google Scholar] [CrossRef]
- Wangchuk, P. Therapeutic Applications of Natural Products in Herbal Medicines, Biodiscovery Programs, and Biomedicine. J. Biol. Act. Prod. Nat. 2018, 8, 1426495. [Google Scholar] [CrossRef]
- Nasim, N.; Sandeep, I.S.; Mohanty, S. Plant-derived natural products for drug discovery: Current approaches and prospects. Nucl. 2022, 65, 399. [Google Scholar] [CrossRef]
- WHO Global Centre for Traditional Medicine. Available online: https://www.who.int/initiatives/who-global-centre-for-traditional-medicine (accessed on 11 April 2023).
- Liu, K.; Ding, X.; Deng, B.; Chen, W. Isolation and characterization of endophytic taxol-producing fungi from Taxus chinensis. J. Ind. Microbiol. Biotechnol. 2009, 36, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Somjaipeng, S.; Medina, A.; Kwaśna, H.; Ortiz, J.O.; Magan, N. Isolation, identification, and ecology of growth and taxol production by an endophytic strain of Paraconiothyrium variabile from English yew trees (Taxus baccata). Fungal Biol. 2015, 119, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Drożdżyński, P. Biosurfaktanty Otrzymywane z Endofitycznych Bakterii Wyizolowanych z Chelidonium majus L.—Produkcja i Zastosowanie. 2021. Available online: https://politechnikalodzka.ssdip.bip.gov.pl/articles/pdf/277859 (accessed on 27 February 2024).
- Zouari Bouassida, K.; Bardaa, S.; Khimiri, M.; Rebaii, T.; Tounsi, S.; Jlaiel, L.; Trigui, M. Exploring the Urtica dioica Leaves Hemostatic and Wound-Healing Potential. Biomed. Res. Int. 2017, 2017, 1047523. [Google Scholar] [CrossRef] [PubMed]
- Naoufal, D.; Amine, H.; Ilham, B.; Khadija, O. Isolation and Biochemical Characterisation of Endophytic Bacillus spp from Urtica dioica and Study of Their Antagonistic Effect against Phytopathogens. Annu. Res. Rev. Biol. 2018, 28, 1–7. [Google Scholar] [CrossRef]
- Chevallier, A. Encyclopedia of Herbal Medicine. Available online: https://www.dk.com/uk/book/9780241593370-encyclopedia-of-herbal-medicine-new-edition/ (accessed on 4 October 2023).
- Tello, S.A.; Silva-Flores, P.; Agerer, R.; Halbwachs, H.; Beck, A.; Peršoh, D. Hygrocybe virginea is a systemic endophyte of Plantago lanceolata. Mycol. Prog. 2014, 13, 471–475. [Google Scholar] [CrossRef]
- Bhukta, P.; Rath, D.; Pattnaik, G.; Kar, B.; Ranajit, S.K. A Comprehensive Review on Promising Phytopharmacological Applications of Chamomile Flower. Asian J. Chem. 2021, 33, 2864–2870. [Google Scholar] [CrossRef]
- Presibella, M.M.; De Biaggi Villas-Bôas, L.; da Silva Belletti, K.M.; de Moraes Santos, C.A.; Weffort-Santos, A.M. Comparison of Chemical Constituents of Chamomilla recutita (L.) Rauschert Essential Oil and its Anti-Chemotactic Activity. Braz. Arch. Biol. Technol. 2006, 49, 717–724. [Google Scholar] [CrossRef]
- Petronilho, S.; Maraschin, M.; Coimbra, M.A.; Rocha, S.M. In vitro and in vivo studies of natural products: A challenge for their valuation. The case study of chamomile (Matricaria recutita L.). Ind. Crop. Prod. 2012, 40, 1–12. [Google Scholar] [CrossRef]
- Köberl, M.; Schmidt, R.; Ramadan, E.M.; Bauer, R.; Berg, G. The microbiome of medicinal plants: Diversity and importance for plant growth, quality, and health. Front. Res. Found. 2013, 4, 400. [Google Scholar] [CrossRef]
- Schmidt, R.; Köberl, M.; Mostafa, A.; Ramadan, E.M.; Monschein, M.; Jensen, K.B.; Bauer, R.; Berg, G. Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 2014, 5, 00064. [Google Scholar] [CrossRef]
- Köberl, M.; White, R.A., III; Erschen, S.; Spanberger, N.; El-Arabi, T.F.; Jansson, J.K.; Berg, G. Complete Genome Sequence of Bacillus amyloliquefaciens Strain Co1-6, a Plant Growth-Promoting Rhizobacterium of Calendula officinalis. Genome Announc. 2015, 3, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Dallio, M.; Loguercio, C. Silymarin/Silybin and chronic liver disease: A marriage of many years. Molecules 2017, 22, 191. [Google Scholar] [CrossRef]
- Schmidt, A.G.H.H.-J. Silymarin as Supportive Treatment in Liver Diseases: A Narrative Review. Adv. Ther. 2020, 37, 1279–1301. [Google Scholar] [CrossRef]
- Saller, R.; Brignoli, R.; Melzer, J.; Meier, R. An updated systematic review with meta-analysis for the clinical evidence of silymarin. Forsch. Komplementärmedizin/Res. Complement. Med. 2008, 15, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Anwar, Y.; Khan, S.U.; Ullah, I.; Hemeg, H.A.; Ashamrani, R.; Al-sulami, N.; Ghazi Alniami, E.; Hashem Alqethami, M.; Ullah, A. Molecular Identification of Endophytic Bacteria from Silybum marianum and Their Effect on Brassica napus Growth under Heavy Metal Stress. Sustainability 2023, 15, 3126. [Google Scholar] [CrossRef]
- Juyal, P.; Shrivastava, V.; Mathur, A. Evaluation of Antimicrobial Potential of Endophytic Fractions of Mentha piperita against Pathogenic Microbes. Int. J. Dev. Res. 2017, 7, 14120–14124. Available online: http://www.journalijdr.com (accessed on 5 May 2024).
- Shokhiddinova, M.N.; Normurodova, Q.T. Isolation of Endophytic Bacteria from Medicinal Plants and Screening for Their Enzyme Production Ability. Appl. Microbiol. Open Access 2024, 9, 1000281. [Google Scholar] [CrossRef]
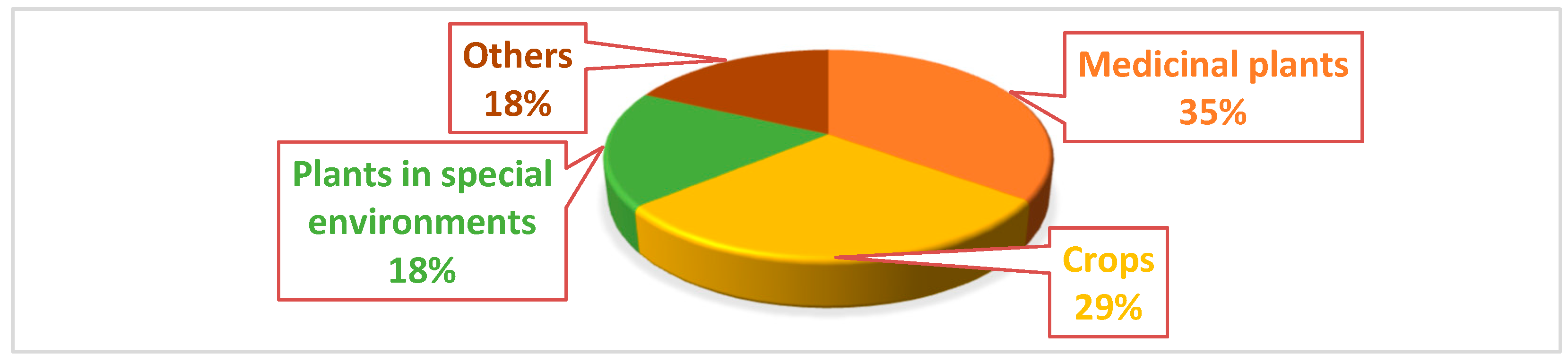
| No. | Method of Plant Material Sterilization | Plant or Plants Part Example | Endophytes Isolated | Reference |
|---|---|---|---|---|
| Single compound approach | ||||
| 1 | EtOH 70–99% (different times) | Nepenthes species; Anemone nemorosa, Ranunculus ficaria, Vaccinium oxycoccus, Sambucus nigra | Acremonium sp. Libertella heveae | [53,54,55] |
| 2 | NaOCl 2–30% (up to 30 min) | leaves stems and roots Pistacia atlantica L.; Simmondsia chinensis | Total of 61 endophytes, with 10 belonging to genus Pseudomonas, Stenotrophomonas, Bacillus, Pantoea and Serratia; Bacillus sp., Methylobacterium aminovorans, Oceanobacillus kimchi, Rhodococcus pyridinivorans, and Streptomyces sp. | [56,57] |
| 3 | 0.1% HgCl2 (1–10 min) | Ziziphus nummularia | Microsporum gypseum, Aspergillus fumigatus, Aspergillus calidoustus, Penicillium viridicatum, Trichophyton tonsurans, and Penicillium marneffei | [58] |
| 4 | Surfactants | Leaves, stems, and roots | [59] | |
| 5 | Tween 20 0.01–10% | Leaves, stems, and roots | 127 Endophytic fungi | [60] |
| 6 | Tween 80 0.1% | Fruits | [61] | |
| 7 | Teepol 5% | Leaves, stems, and roots | 228 Isolates representing at least 19 genera of actinobacteria | [62] |
| 8 | Triton X-100 0.05–0.1% | Leaves, stems, and roots | 116 endophytic fungi | [63,64,65] |
| 10 | Hydrogen peroxide 3–90% | Leaves, stems, and roots | [59,66] | |
| Combined approach | ||||
| 11 | 2% NaOCl + 0.1% Tween 20 | Roots (tomato) | Rhizobium, Bacillus, Microbacterium, Enterobacter species | [67,68] |
| 12 | 70% EtOH-3.125% NaOCl | Psidium guajava and Ziziphus mauritiana | 7 Endophytic actinomycetes | [53] |
| 13 | 75% EtOH (2 min) and 1.5%NaOCl (3 min) | Brassica olerocea, B. rapa, and Raphanus sativus | 4178 Endophytic fungal isolates belonging to 51 different genera | [69] |
| 14 | 70% EtOH (1 min), 2.5% NaOCl (3 min); 70% EtOH (1 min), 2.5% NaOCl (1 min), and 70% EtOH (30 s) | Roots and leaves of Salvia abrotanoides | 56 Endophytes | [70] |
| 15 | 75% EtOH (2 min), 2% NaOCl (3 min), and 75% EtOH (30 s) | Gnetum gnemon | Staphylococcus warneri, Solibacillus isronensis, Bacillus megaterium, Caballeronia glebae; Bacillus licheniformis, Bacillus velezensis, and Bacillus atrophaeus | [71] |
| 16 | 75% EtOH (1 min), 1% HgCl2 (4 min), and 75% EtOH (1 min) | Lycoris radiata | Total of 188 bacterial endophytes | [72] |
| 17 | 70% EtOH (1 min), 2.5% NaOCl (2 min), and 70% EtOH (1 min) | Stems, leaves, and capsules of Spiranthes spiralis, Serapiasvomeracea, and Neottia ovata | 50 bacteria belonging to genera Sphingomonas, Microbacterium, Pantoea, Staphylococcus, Pseudomonas, Bacillus, and Streptomyces | [73] |
| 18 | 75% EtOH (1 min), 1% NaOCl (3 min), and 75% EtOH (30 s) | Artemisia nilagirica | Arthrobacter sp., Bacillus sp., Burkholderia sp., Pseudomonas sp., Psychrobacter sp., Serratia sp., Microbacterium sp., Enterobacter sp., Chromobacterium violaceum, and Kosakonia cowanii | [74] |
| 19 | 75% EtOH (1 min) and 5% NaOCl (8 min) | Lycium ruthenicum | 109 Endophytic bacteria with 36 genera | [65,68] |
| 20 | Cycloheximide (100 mg/L) and nalidixic acid (20 mg/L) (1 min), 5% NaOCl 5 min), 2.5% Na2S2O3 (10 min), and 10% NaHCO3 (10 min) | Glycine max | 70 Endophytic actinobacteria belonging to 14 genera | [75] |
| 21 | 70% EtOH (3 min), 8% NaOCl (4–5 min), 2.5% Na2S2O3 (10 min), and 70% EtOH (1 min) | Camellia sinensis | 46 Actinobacteria belonging to families Streptomycetaceae, Thermomonosporaceae, Camellia sinensis, Nocardioidaceae, Microbacteriaceae, Dermatophilaceae, Nocardiopsaceae, Nocardiaceae, Mycobacteriaceae, Dermacoccaceae, Micromonosporaceae, and Pseudonocardiaceae | [76] |
| 22 | 70% EtOH (1 min), 1.2% NaOCl (15 min), and 75% EtOH (30 s) | Oryza sativa | 87 Endophytic bacteria | [77] |
| 23 | 2.5% NaOCl (2 min), and 75% EtOH (30 s) | Pinus cotorta | 77 Endophytic diazotrophs Pseudomonas, Bacillus, Paenibacillus, and Rhizobium | [78] |
| 24 | Surfactant-75% EtOH (10 min), 5.25% NaOCl (10 min), and 70% EtOH (2 min) | Phoenix dactylifera L. | 14 Endophytic bacteria | [79] |
| 25 | 5% NaOCl (4–10 min), 2.5% Na2S2O3 (10 min), and 75% EtOH and 10% NaHCO3 (10 min) | 12 Forest trees | 2174 Actinobacteria | [80] |
| 26 | 0.1% Tween 20 and 70% EtOH (3 min), 0.4% NaOCl (1 min), and 70% EtOH (2 min) | 7 Medicinal plants | 13 Streptomyces sp., 2 Microbacterium sp., 1 Leifsonia xyli, 1 Brevibacterium sp., 1 Actinomycete | [81] |
| 30 | 0.01% Tween 20 (1 min), 2.5% NaOCl (2.5 min), 2.5% Na2S2O3 (5 min), 75% EtOH (2.5 min), and 10% NaHCO3 (5 min) | Dioscorea zingiberensis | 123 Strains of endophytic fungi, most abundant genera | [77] |
| No | Host Plant | Endophytic Bacteria | Compound/-s | Activity | Reference |
|---|---|---|---|---|---|
| 1 | Achillea fragrantissima | Streptomyces sp., Nocardioides sp., Kitasatosporia sp., Kibdelosporangium sp. | chitinase, siderophores | enzymatical, antifungal | [98] |
| 2 | Achillea millefolium (yarrow) | Bacillus safensis | volatile metabolites (butanal, 3-methyl-, 2-heptanone, 6-methyl-5-methylene-, hydrogen azide, propene, 2-butene, 6-oxabicyclo3.1.0 hexane) | antifungal | [99] |
| 3 | Alkanna tinctoria (dyer’s alkanet) | Pseudomonas sp., Bacillus sp. | IAA, ACC deaminase, siderophore; pectinase, ligninase | plant growth promotion; enzymatical | [100] |
| 4 | Allium fistulosum (spring onion) | Streptomyces sp. TP-AO569 | fistupyrone | spore germination inhibition | [101] |
| 5 | Allium tuberosum (garlic chives) | Streptomyces sp. TP-A0595 | 6-prenylidole | antifungal | [102] |
| 6 | Arctium lappa (greater burdock) | Bacillus sp., Pantoea sp., Microbacterium sp., Pseudomonas sp. | IAA, ACC deaminase, siderophore, hydrolase | plant growth promotion | [103] |
| 7 | Armoracia rusticana (horseradish) | Serratia ficaria | siderophores, lipase, protease, chirinase | biocontrol, enzymatic, plant growth promotion | [104] |
| 8 | Arnica montana (mountain arnica) | Streptomyces sp. | glutarimide antibiotics (cycloheximide, actiphenol), diketopiperazines (cyclo-prolyl-valyl, cyclo-prolyl-isoleucyl, cyclo-prolyl-leucyl, cyclo-prolyl-phenylalanyl) | antibiotic, cytotoxic | [105] |
| 9 | Artemisia vulgaris (common mugworts) | Bacillus pumilus, Bacillus safensis | volatile metabolites (butanal, 3-methyl-, 2-heptanone, 6-methyl-5-methylene-, hydrogen azide, propene, 2-butene, 6-oxabicyclohexane) | antifungal | [99] |
| 10 | Atropa belladonna (deadly nightshade) | Streptomyces sp. AB100 | piperazic-acid-containing peptides | antibiotic | [106] |
| 11 | Calendula arvensis (field marigold) | Pseudomonas brassicacearum | - | biocontrol, plant growth promotion | [107] |
| 12 | Calendula officinalis (medicinal calendula) | Pseudomonas putida | HCN, siderophores, lipase, protease, chitinase | biocontrol, enzymatic, plant growth promotion | [104] |
| 13 | Matricaria chamomilla L., Calendula officinalis L., and Solanum distichum Schumach. and Thonn | Bacillus subtilis subsp. subtilis, Bacillus subtilis subsp. spizizenii | - | antibacterial, antifungal, nematicidal | [108] |
| 14 | Iris pseudacorus L. | Pseudomonas rhizosphaereae FST5 | IAA, ACC deaminase, HCN, chitinase, protease, glucanase, lipase | antifungal, plant growth promotion | [109] |
| 15 | Chelidonium majus (greater celandine) | Bacillus velezensis | IAA, siderophores, iturin B, iturin D, fengycin, surfactin, cellulase | biocontrol, plant growth promotion | [110] |
| 16 | Chelidonium majus (greater celandine) | Bacillus pumilus 2A | glycolipid | emulsyfing (biosurfactant) | [111] |
| 17 | Chelidonium majus (greater celandine) | Bacillus amyloliquefaciens 30B, Erwinia persicinia 2–5b | IAA | plant growth promotion | [112] |
| 18 | Echinacea purpurea (purple coneflower) | Bacillus thuringensis, Bacillus amyloliquefaciens | - | antifungal | [113] |
| 19 | Echinacea purpurea (purple coneflower) | Rheinheimera sp. EpRS3 | lipases, phospholipase, protease | antibacterial, antimicrobial | [114] |
| 20 | Elymus repens (couch grass) | Pseudomonas azotoformans | IAA | plant growth promotion | [115] |
| 21 | Equisetum arvense (common horsetail) | Psychrobacillus insolitus, Curtobacterium oceanosedimentum | - | anticandidal | [116] |
| 22 | Euphorbia helioscopia (sun spurge) | Comamonas koreensis, Stenotrophomonas maltophilia, Rhizobium sp., Brevundimonas sp. | IAA-like compounds, siderophores | plant growth promotion | [117] |
| 23 | Euphorbia helioscopia (sun spurge) | Bacillus cereus, Bacillus amyloliquefaciens | - | biocontrol, plant growth promotion | [107] |
| 24 | Euphorbia peplus (petty spurge) | Pseudomonas brassicacearum | - | biocontrol, plant growth promotion | [107] |
| 25 | Foeniculum vulgare (fennel) | Klebsiella pneumoniae | siderophores, lipase, protease, chitinase | biocontrol, enzymatic, plant growth promotion | [104] |
| 26 | Hypericum perforatum (St. John’s wort) | Achromobacter sp., Erwinia persicina, Stenotrophomonas sp. | IAA, HCN, cellulase, protease, beta-1,3-glucanase | biocontrol, plant growth promotion, antifungal | [118] |
| 27 | Iris pseudacours (pale yellow iris) | Pseudomonas azotoformans | HCN, siderophores, lipase, protease | biocontrol, enzymatic, plant growth promotion | [104] |
| 28 | Matricaria chamomilla L., Calendula officinalis L., and Solanum distichum Schumach. and Thonn | Pseudomonas gessardii HRT18 | IAA, HCN, siderophores, lipase, protease, chitinase, glucanase | biocontrol, enzymatic, plant growth promotion | [109] |
| 29 | Lavandula dentata (fringed lavender) | Pseudomonas sp., Bacillus sp. | IAA, HCN, siderophores, lipase, protease, cellulase, pectinase | plant growth promotion | [11] |
| 30 | Lotus corniculatus (bird’s foot trefoil) | Serratia plymuthica, Pseudomonas mandelii, Tsukamurella pulmonis | IAA, HCN, siderophores, cellulase | plant growth promotion | [119] |
| 31 | Matricaria chamomilla (German chamomile) | Bacillus pumilus | lipopeptides | antifungal | [99] |
| 32 | Calendula officinalis | Bacillus mojavensis, Bacillus subtilis subsp. subtilis, Bacillus subtilis subsp. spizizenii, Bacillus endophyticus, Paenibacillus brasilensis, Paenibacillus polymyxa, Lysobacter enzymogenes | - | antibacterial, antifungal, nematicidal | [108] |
| 33 | Mentha longifolia (wild mint) | Bacillus sp., Bacillus aryabhattai, Bacillus pumilus, Bacillus megaterium, Bacillus toyonensis | IAA, ACC deaminase, siderophores | plant growth promotion | [120] |
| 34 | Mentha spicata (spearmint) | Bacillus anthracis, Bacillus toyonensis | cellulase, xylanase, amylase, pectinase | enzymatical | [121] |
| 35 | Polygonum cuspidatum | Bacillus safensis | volatile metabolites (butanal, 3-methyl-, 2-heptanone, 6-methyl-5-methylene-, hydrogen azide, propene, 2-butene, 6-oxabicyclo3.1.0 hexane) | antifungal | [99] |
| 36 | Oenothera biennis (common evening primrose) | Rhodococcus erythropolis, Rhizobium sp. | IAA, HCN, cellulase | plant growth promotion | [119] |
| 37 | Plantago lanceolata (ribwort plantain) | Pseudomonas brassicacearum, Bacillus methylotrophicus | - | biocontrol, plant growth promotion | [107] |
| 38 | Calendula officinalis | Bacillus halotolerans Cal.l.30 | surfactin, iturin, fengycin, bacillaene, bacillibactin, FAS-PKS, subtilosin A, bacilysin | antibacterial, antifungal, nematicidal | [122] |
| 39 | Peperomia dindygulensis | Streptomyces sp. YINM00001 | cyclohexiamide, dinactin, warkmycin, anthramycin, alkylresorcinol, lanthipeptide, melanin, ectoine, geosmin | antibacterial, antifungal | [123] |
| 40 | Origanum vulgare | Bacillus sp. | paeninodin, terpene, paenilarvins | biocontrol | [124] |
| Paenibacillus sp. | polymyxin, paenicidin A | antimicrobial | |||
| 41 | Mikania micrantha | Sphingomonas paucimobiliz | Zeaxanthin | antioxidant | [105] |
| Micrococcus yunnanensis | microansamycin, stenothricin | biocontrol | |||
| 42 | Mimosa pudica | Staphylococcus caprae | Aureusimine, staphyloferrin A | Plant growth promotion | |
| Neobacillus drentensis | Fengycin | biocontrol | |||
| Priestia megaterium | Surfactin, bacitracin, carotenoid | biocontrol | |||
| 43 | Millettia pachycarpa Benth | Paenibacillus peoriae IBSD35 | NRPSs, Fusaricidin synthetase, paenibacterin, gramicidin synthase | Biocontrol, antibiotic | [114] |
| 44 | Panicum turgidum | Cellulosimicrobium sp. JZ28 | alkylresorcinol | Biocontrol | [125] |
| 45 | Brassica napus L. | Pseudomonas fluorescens BRZ63 | alginate, LPS, siderophore, bacterioferrin, tryptophan, PQQ cofactor | biocontrol, enzymatic, plant growth promotion | [126] |
| 46 | Suaeda fruticosa, Suaeda mollis, Mesembryanthmum nodiflorum, Arthrocnemum indicum | Bacillus albus strains | petrobactin, bacitracin, thuricin | Biocontrol | [127] |
| 47 | Bacopa monnieri | Bacillus sp. LCF1 | surfactin, iturin, fengycin, type I PKS | Biocontrol | [128] |
| 48 | Arnica montana L. | Streptomyces sp. | siderophore, ectoine, terpene, bacteriocin, butyrolactone, lantipeptide, melanin, NRPS, PKS, cyclohexiaminde | biocontrol, enzymatic, plant growth promotion | [128] |
| 49 | Echinacea purpurea | Rheinheimera sp. RS3 | Resorcinol, lantipeptide, hserlactone, bacteriocin, NRPS | Antimycobacterial potential, biocontrol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drożdżyński, P.; Rutkowska, N.; Rodziewicz, M.; Marchut-Mikołajczyk, O. Bioactive Compounds Produced by Endophytic Bacteria and Their Plant Hosts—An Insight into the World of Chosen Herbaceous Ruderal Plants in Central Europe. Molecules 2024, 29, 4456. https://doi.org/10.3390/molecules29184456
Drożdżyński P, Rutkowska N, Rodziewicz M, Marchut-Mikołajczyk O. Bioactive Compounds Produced by Endophytic Bacteria and Their Plant Hosts—An Insight into the World of Chosen Herbaceous Ruderal Plants in Central Europe. Molecules. 2024; 29(18):4456. https://doi.org/10.3390/molecules29184456
Chicago/Turabian StyleDrożdżyński, Piotr, Natalia Rutkowska, Magdalena Rodziewicz, and Olga Marchut-Mikołajczyk. 2024. "Bioactive Compounds Produced by Endophytic Bacteria and Their Plant Hosts—An Insight into the World of Chosen Herbaceous Ruderal Plants in Central Europe" Molecules 29, no. 18: 4456. https://doi.org/10.3390/molecules29184456










