Insights into the Molecular Structure, Stability, and Biological Significance of Non-Canonical DNA Forms, with a Focus on G-Quadruplexes and i-Motifs
Abstract
:1. Introduction
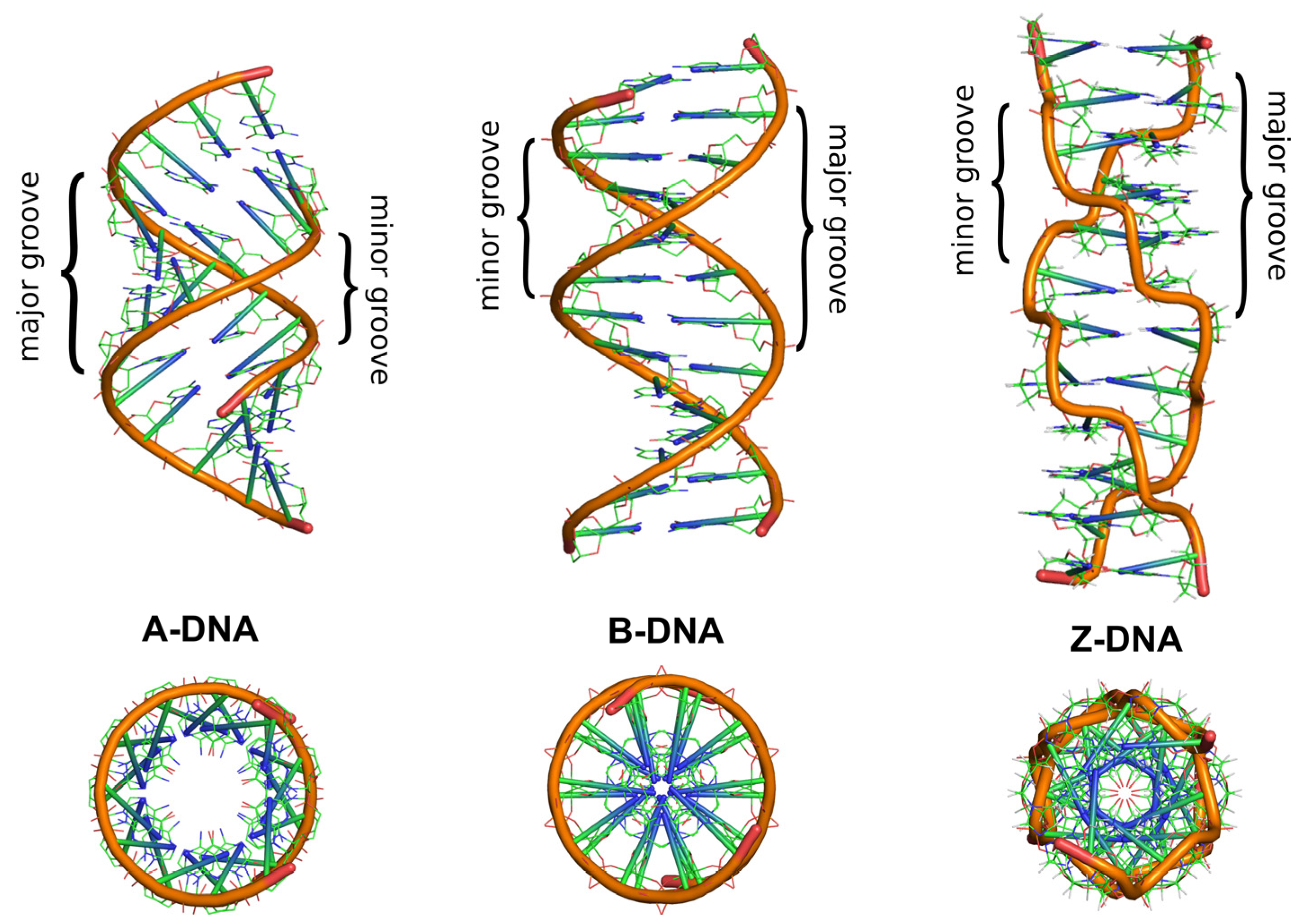
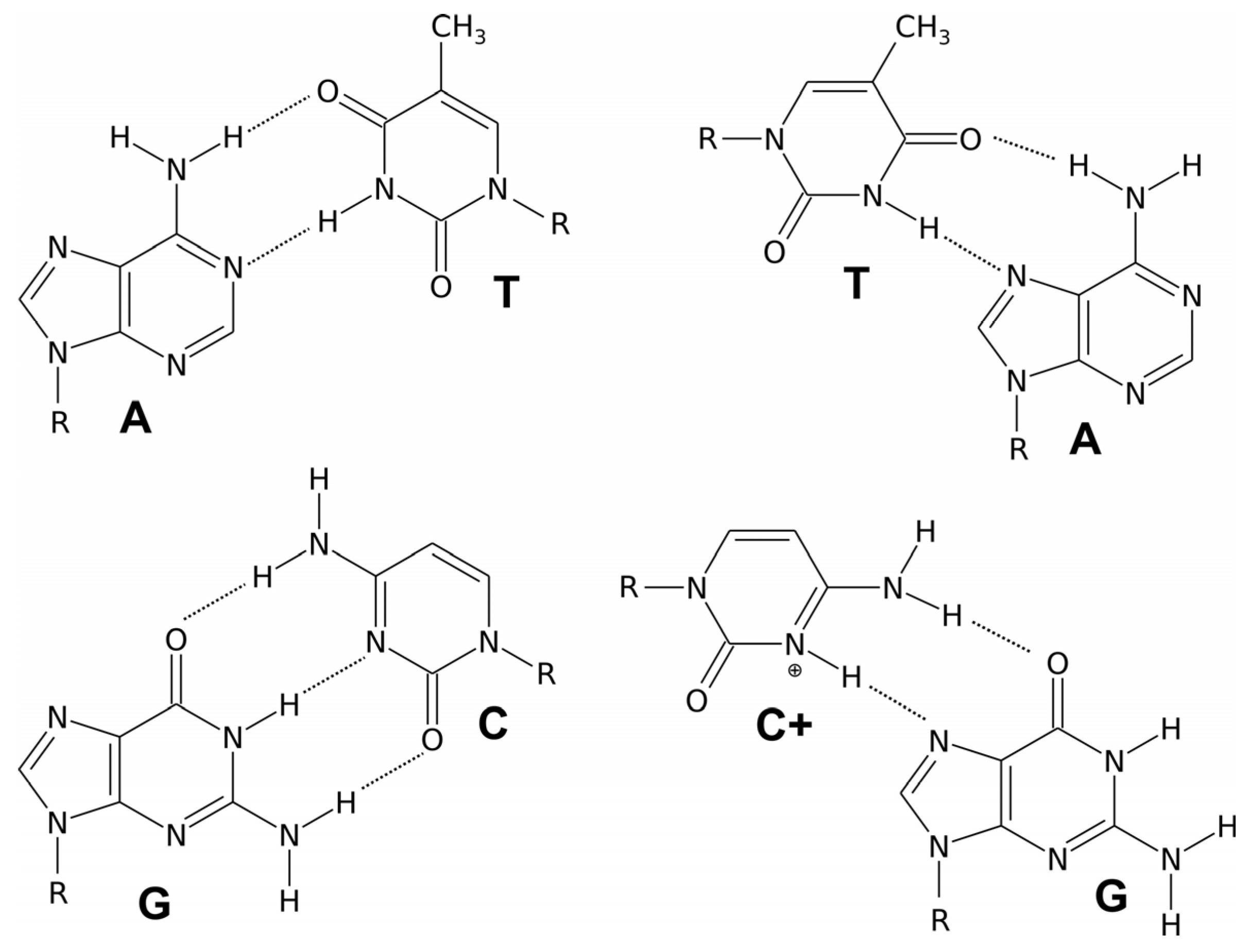
2. Double-Helical DNA Structures
3. Triple-Helical DNA Structures
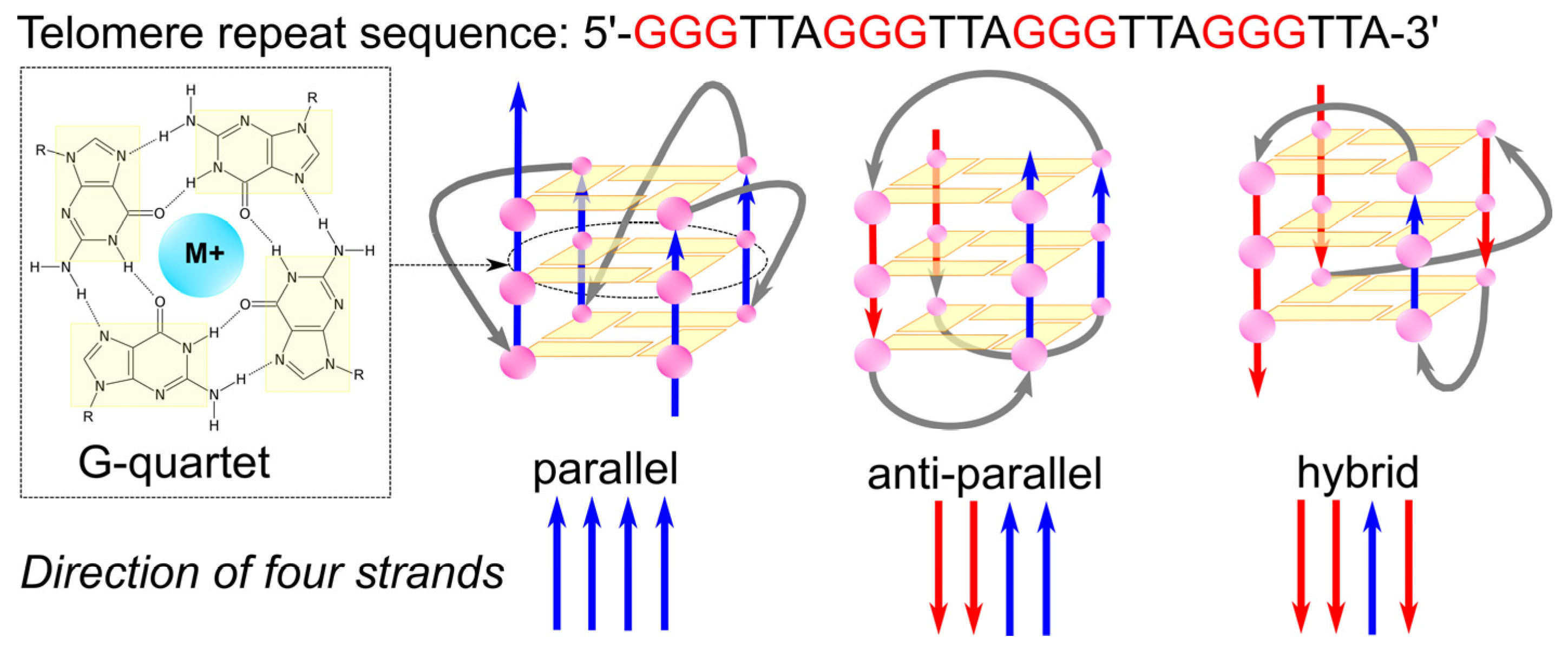
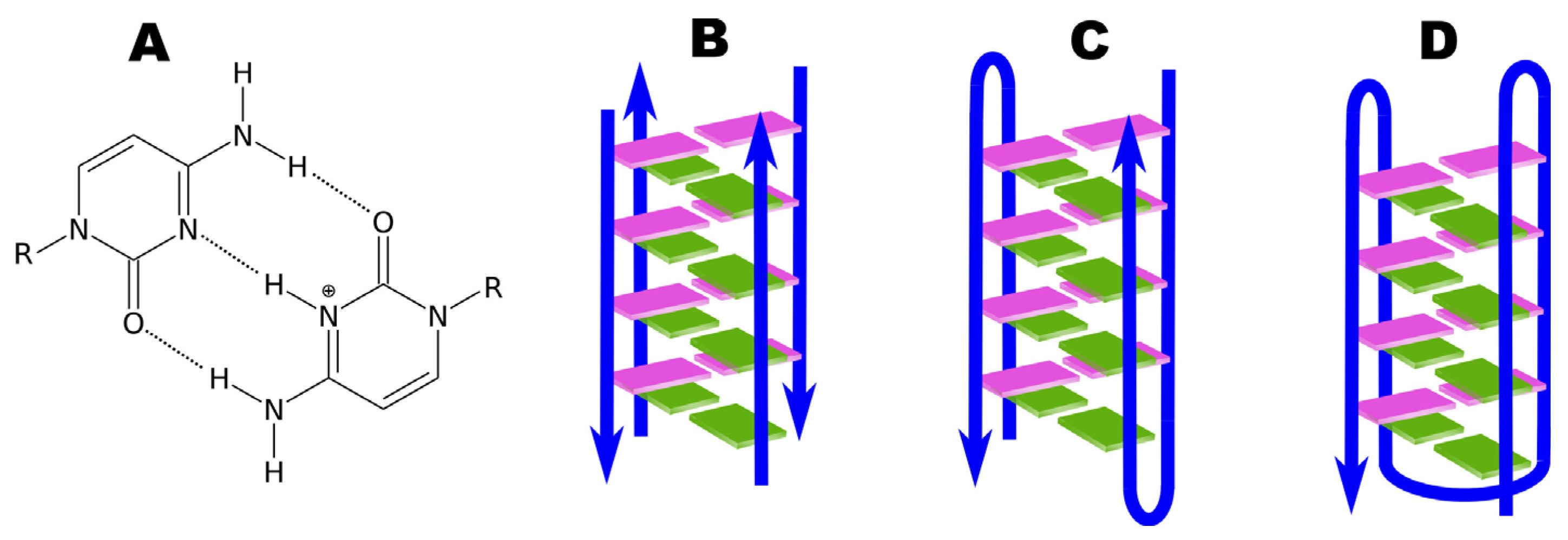
4. Four-Stranded DNA Structures: G-Quadruplexes and i-Motifs
5. Ligands Stabilizing Four-Stranded Non-Canonical Forms of DNA
5.1. G-Quadruplex
5.2. i-Motif
6. Computer Simulations of Four-Stranded Non-Canonical DNA Forms
6.1. G-Quadruplexes
- Ion Coordination and Stabilization: The role of monovalent cations (K+ and Na+) is pivotal in stabilizing the G-quadruplex structures. Simulations have shown that potassium ions, due to their size and coordination properties, fit perfectly within the central channel of G-quadruplexes, stabilizing the G-tetrads through electrostatic interactions [206,207]. Sodium ions, while also stabilizing, do so less effectively compared to potassium, often leading to different conformational preferences in the G-quadruplex structure [201,208,209]. Kinetic analysis based on Markov modeling showed that presence of Na+ modestly enhances an antiparallel G-quadruplex topology, while K+ drives G-quadruplex into a parallel/hybrid topology with much higher affinity than Na+ does [210].
- Folding Pathways and Kinetics: Simulations have provided insights into the folding pathways of G-quadruplexes, revealing multiple intermediate states that the DNA strands can adopt before forming the stable G-quadruplex structure. In general, the folding of G-quadruplexes is best described by a kinetic partitioning (KP) mechanism. KP involves competition between at least two (and often many) well-separated and structurally distinct conformational ensembles. The KP folding landscape contrasts with the funneled landscape, containing deep competing free-energy minima (alternative folds or conformational basins) separated by large free-energy barriers. Only a fraction of molecules fold directly to the native basin, which is most populated at thermodynamic equilibrium. Other molecules initially fold into competing (non-native) basins, becoming trapped in different basins. Thermodynamic equilibrium is reached after numerous misfolding–unfolding events, leading to the equilibrium population of all basins. Therefore, the whole process is slow. Human telomeric G-quadruplex sequences can exhibit multiple folds at thermodynamic equilibrium, with other basins transiently populated during folding. The relative stabilities of different basins can be significantly influenced by the environment [211]. The MD simulations indicate that the immense complexity of the G-quadruplex folding landscape is linked to the ability of many G-quadruplex-folding sequences to adopt multiple alternative structures with different patterns of anti and syn guanosines, which, once formed, have long lifetimes. If these structures appear during folding but are absent in the final thermodynamic equilibrium, detecting and structurally resolving them becomes very challenging [211,212,213]. Bian et al. [214] employed a hybrid atomistic structure-based model to investigate the folding dynamics of the human telomeric DNA G-quadruplex. This model integrates structural information from three known G-quadruplex topologies: hybrid 1, hybrid 2, and chair-type conformations. The model was validated by its ability to replicate experimental observations, specifically that the hybrid-1 conformation is the major fold while hybrid 2 is more kinetically accessible. A three-step mechanism was identified for the formation of the hybrid 1 conformation, whereas the hybrid 2 and chair-type conformations followed a two-step mechanism. The presence of inappropriate syn/anti guanine nucleotide combinations was found to slow down the folding process significantly. In a recent study, Kim et al. [213] proposed a folding scheme for the human telomeric G-quadruplex using state-of-the-art enhanced sampling molecular dynamics simulations at the all-atom level. As illustrated in Figure 8, the G-quadruplex folding process begins with the formation of a single-hairpin structure, followed by the formation of double hairpins. These double hairpins then proceed along distinct folding pathways, leading to various G-quadruplex topologies, including antiparallel chair, antiparallel basket, hybrids 1 and 2, and parallel propeller forms. Additionally, three-triad and two-tetrad structures with antiparallel backbone alignment act as crucial intermediates, facilitating the folding process and transitions between different G-quadruplex structures. This computational study also demonstrated that the structural ensemble and ion capture by human telomeric DNA dramatically respond to temperature increases.
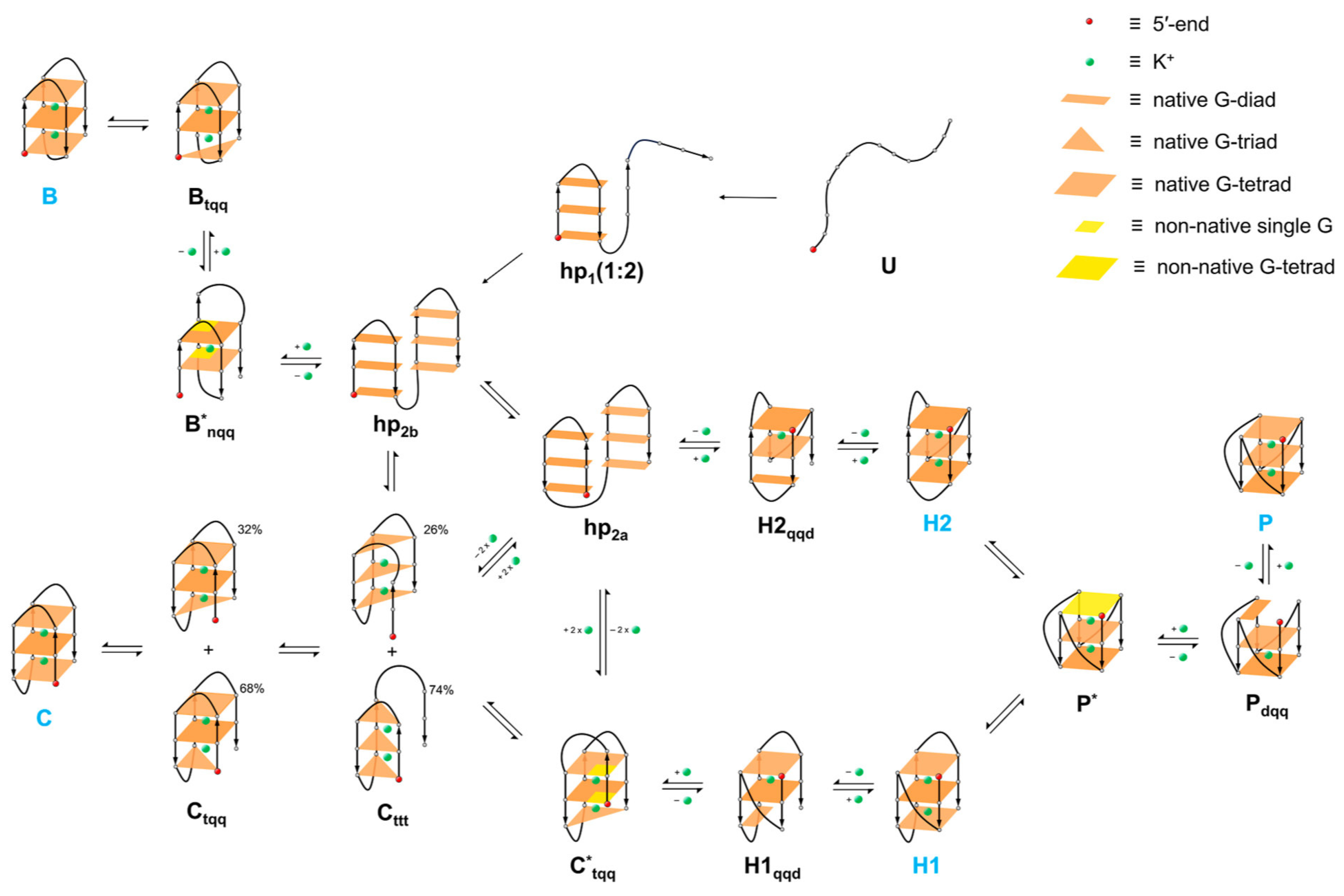
- Loop Dynamics and Conformational Flexibility: The dynamics and flexibility of the loops connecting the G-tetrads are crucial for the overall stability and folding of G-quadruplexes [215]. Simulations have shown that loop length and sequence composition can dramatically affect the folding kinetics and stability of the resulting structure [216]. The simulation results by Islam et al. [217] suggest that the loops may exist as a dynamic continuum of interconverting substates, which would be difficult to fully capture by available experimental methods. Additionally, they demonstrated that long simulations are needed to sufficiently exhaustively characterize quadruplex DNA loop dynamics without visible bias from the starting structure. One of the most interesting structural observations was the end capping of a quadruplex with the terminal adenine base. Markov-state modeling was employed to understand the trends of structural transitions in the propeller loops. Transition rates estimated by transition path theory indicated that loop interconversions occur on microsecond to dozens of microseconds time scales. Using the bsc0 AMBER force field, simulations visualized all the main conformational substates on the landscape of the TTA propeller loops [218]. Studies of decomposition thermodynamics indicated that the G-tetrad is strongly stabilized by interactions involving the sugar–phosphate backbone and TTA loops. The energetics of guanine association alone is not the decisive factor [219].
- Hydration and Solvent Effects: The role of water molecules and hydration shells around G-quadruplexes has been extensively studied using MD simulations. These studies have highlighted the importance of water-mediated interactions in stabilizing the G-tetrads and influencing the overall conformation of the G-quadruplex. Chowdhury and Bansal found that the guanine quadruplex is stable, even in the absence of coordinated cations. Water molecules can occupy the empty coordination sites in this situation. Sodium ions can enter a preformed quadruplex through the ends and travel within the quadruplex channel without significantly distorting the G-tetrad geometry. Meanwhile, water molecules can exit the channel through the ends as well as through the grooves [206]. Additionally, the presence of water molecules is essential for the accurate representation of the folding landscapes of G-quadruplexes in simulations. Hydration shells around the DNA provide a realistic environment that affects the energy barriers and the pathways of folding and unfolding processes. Long simulations are necessary to capture the exhaustive dynamics of these hydration effects without bias from the starting structures [217]. These findings underscore the intricate interplay between G-quadruplexes and their solvent environment, highlighting the necessity of considering solvent effects in computational studies of these structures. Understanding these interactions is crucial for accurate modeling of G-quadruplex stability, folding mechanisms, and their biological functions [220].
- Ligands Stabilizing G-quadruplexes: Identifying and designing ligands that selectively bind and stabilize G-quadruplexes is of great interest, particularly for therapeutic applications in oncology. Ligands such as telomestatin and various small molecules have been shown to preferentially stabilize G-quadruplex structures over duplex DNA, thereby inhibiting the activity of telomerase and certain oncogenes. Docking studies, ligand-based methods, especially QSAR (Quantitative Structure–Activity Relationships), pharmacophore models, and MD simulations have been pivotal in understanding the binding modes and affinities of these ligands [221,222]. For instance, Ramos et al. [200] investigated a diketopyrrolo[3,4-c]pyrrole derivative and found that it binds to G-quadruplexes through various modes, significantly stabilizing these structures and inhibiting oncogene promoter activity. Mulliri et al. [223] studied substituted pyrazolo[1,2-a]benzo[1,2,3,4]tetrazine-3-one derivatives as G-quadruplex stabilizing/destabilizing ligands. The MD results in this study were particularly important when considering that the docking study indicated that both the stabilizing and destabilizing compounds display a similar negative binding free energy, while the MD simulations discriminated the stabilizing/destabilizing activity of the ligands.
6.2. i-Motifs
7. Summary
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berg, J.M.; Tymoczko, J.L.; Gatto, G.J.; Stryer, L. Biochemistry, 9th ed.; Macmillan International, Higher Education: New York, NY, USA, 2019; ISBN 978-1-319-11465-7. [Google Scholar]
- Allison, L.A. Fundamental Molecular Biology, 3rd ed.; Wiley Blackwell: Hoboken, NJ, USA; Chichester, UK, 2021; ISBN 978-1-119-15629-1. [Google Scholar]
- Neidle, S. (Ed.) Oxford Handbook of Nucleic Acid Structure; Oxford University Press: Oxford, UK; New York, NY, USA, 1999; ISBN 978-0-19-850038-4. [Google Scholar]
- Vicens, Q.; Kieft, J.S. Thoughts on How to Think (and Talk) about RNA Structure. Proc. Natl. Acad. Sci. USA 2022, 119, e2112677119. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.C. (Ed.) Molecular Biology, 3rd ed.; BIOS instant notes; Taylor & Francis: New York, NY, USA, 2005; ISBN 978-0-415-35167-6. [Google Scholar]
- Phan, A.T.; Mergny, J.-L. Human Telomeric DNA: G-Quadruplex, i-Motif and Watson–Crick Double Helix. Nucleic Acids Res. 2002, 30, 4618–4625. [Google Scholar] [CrossRef]
- Barczak, W.; Rozwadowska, N.; Romaniuk, A.; Lipińska, N.; Lisiak, N.; Grodecka-Gazdecka, S.; Książek, K.; Rubiś, B. Telomere Length Assessment in Leukocytes Presents Potential Diagnostic Value in Patients with Breast Cancer. Oncol. Lett. 2016, 11, 2305–2309. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-H.; Chen, R.-J. Prevalence of Telomerase Activity in Human Cancer. J. Formos. Med. Assoc. 2011, 110, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.L.; Hirose, Y.; Langmore, J.P. Long G Tails at Both Ends of Human Chromosomes Suggest a C Strand Degradation Mechanism for Telomere Shortening. Cell 1997, 88, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct Evidence for a G-Quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress c- MYC Transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Brooks, T.A.; Kendrick, S.; Hurley, L. Making Sense of G-quadruplex and I-motif Functions in Oncogene Promoters. FEBS J. 2010, 277, 3459–3469. [Google Scholar] [CrossRef]
- Day, H.A.; Pavlou, P.; Waller, Z.A.E. I-Motif DNA: Structure, Stability and Targeting with Ligands. Bioorg. Med. Chem. 2014, 22, 4407–4418. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, J.; Gao, Y.; Pan, W.; Yang, Y.; Li, X.; Chen, L.; Wang, C.; Wang, Y. Emerging Roles of I-Motif in Gene Expression and Disease Treatment. Front. Pharmacol. 2023, 14, 1136251. [Google Scholar] [CrossRef]
- Zanin, I.; Ruggiero, E.; Nicoletto, G.; Lago, S.; Maurizio, I.; Gallina, I.; Richter, S.N. Genome-Wide Mapping of i-Motifs Reveals Their Association with Transcription Regulation in Live Human Cells. Nucleic Acids Res. 2023, 51, 8309–8321. [Google Scholar] [CrossRef]
- Hegyi, H. Enhancer-Promoter Interaction Facilitated by Transiently Forming G-Quadruplexes. Sci. Rep. 2015, 5, 9165. [Google Scholar] [CrossRef] [PubMed]
- Yella, V.R.; Vanaja, A.; Kulandaivelu, U.; Kumar, A. Delving into Eukaryotic Origins of Replication Using DNA Structural Features. ACS Omega 2020, 5, 13601–13611. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Vuong, B.Q.; Vaidyanathan, B.; Lin, J.-Y.; Huang, F.-T.; Chaudhuri, J. Non-Coding RNA Generated Following Lariat Debranching Mediates Targeting of AID to DNA. Cell 2015, 161, 762–773. [Google Scholar] [CrossRef]
- Ghosh, A.; Bansal, M. A Glossary of DNA Structures from A to Z. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Richmond, T.J.; Davey, C.A. The Structure of DNA in the Nucleosome Core. Nature 2003, 423, 145–150. [Google Scholar] [CrossRef]
- Chen, F.Y.-H.; Park, S.; Otomo, H.; Sakashita, S.; Sugiyama, H. Investigation of B-Z Transitions with DNA Oligonucleotides Containing 8-Methylguanine. Artif. DNA PNA XNA 2014, 5, e28226. [Google Scholar] [CrossRef]
- Yang, L.; Wang, S.; Tian, T.; Zhou, X. Advancements in Z-DNA: Development of Inducers and Stabilizers for B to Z Transition. Curr. Med. Chem. 2012, 19, 557–568. [Google Scholar] [CrossRef]
- Sinden, R.R. DNA Structure and Function; Academic Press: San Diego, CA, USA, 1994; ISBN 978-0-12-645750-6. [Google Scholar]
- Zhang, H.; Yu, H.; Ren, J.; Qu, X. Reversible B/Z-DNA Transition under the Low Salt Condition and Non-B-Form PolydApolydT Selectivity by a Cubane-Like Europium-L-Aspartic Acid Complex. Biophys. J. 2006, 90, 3203–3207. [Google Scholar] [CrossRef]
- Marx, J.L. Z-DNA: Still Searching for a Function: Six Years after the Discovery of Z-DNA Questions Remain about Whether It Exists Naturally and What Its Functions Might Be. Science 1985, 230, 794–796. [Google Scholar] [CrossRef]
- Choi, J.; Majima, T. Conformational Changes of Non-B DNA. Chem. Soc. Rev. 2011, 40, 5893. [Google Scholar] [CrossRef]
- Chou, S.-H. Unusual DNA Duplex and Hairpin Motifs. Nucleic Acids Res. 2003, 31, 2461–2474. [Google Scholar] [CrossRef]
- Lee, A.-R.; Kim, N.-H.; Seo, Y.-J.; Choi, S.-R.; Lee, J.-H. Thermodynamic Model for B-Z Transition of DNA Induced by Z-DNA Binding Proteins. Molecules 2018, 23, 2748. [Google Scholar] [CrossRef] [PubMed]
- Zain, R.; Sun, J.-S. Do Natural DNA Triple-Helical Structures Occur and Function in Vivo? CMLS Cell. Mol. Life Sci. 2003, 60, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Glazer, P.M. Triplex DNA: Fundamentals, Advances, and Potential Applications for Gene Therapy. J. Mol. Med. 1997, 75, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G.; Davies, D.R.; Rich, A. Formation of a Three-Stranded Polynucleotide Molecule. J. Am. Chem. Soc. 1957, 79, 2023–2024. [Google Scholar] [CrossRef]
- Frank-Kamenetskii, M.D.; Mirkin, S.M. Triplex DNA Structures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef]
- Dalla Pozza, M.; Abdullrahman, A.; Cardin, C.J.; Gasser, G.; Hall, J.P. Three’s a Crowd—Stabilisation, Structure, and Applications of DNA Triplexes. Chem. Sci. 2022, 13, 10193–10215. [Google Scholar] [CrossRef]
- Nikolova, E.N.; Kim, E.; Wise, A.A.; O’Brien, P.J.; Andricioaei, I.; Al-Hashimi, H.M. Transient Hoogsteen Base Pairs in Canonical Duplex DNA. Nature 2011, 470, 498–502. [Google Scholar] [CrossRef]
- Semenyuk, A.; Darian, E.; Liu, J.; Majumdar, A.; Cuenoud, B.; Miller, P.S.; MacKerell, A.D.; Seidman, M.M. Targeting of an Interrupted Polypurine:Polypyrimidine Sequence in Mammalian Cells by a Triplex-Forming Oligonucleotide Containing a Novel Base Analogue. Biochemistry 2010, 49, 7867–7878. [Google Scholar] [CrossRef]
- Ranasinghe, R.T.; Rusling, D.A.; Powers, V.E.C.; Fox, K.R.; Brown, T. Recognition of CG Inversions in DNA Triple Helices by Methylated 3H-Pyrrolo[2,3-d]Pyrimidin-2(7H)-One Nucleoside Analogues. Chem. Commun. 2005, 20, 2555. [Google Scholar] [CrossRef]
- Rusling, D.A. Four Base Recognition by Triplex-Forming Oligonucleotides at Physiological pH. Nucleic Acids Res. 2005, 33, 3025–3032. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Wang, G.; Vasquez, K.M. DNA Triple Helices: Biological Consequences and Therapeutic Potential. Biochimie 2008, 90, 1117–1130. [Google Scholar] [CrossRef]
- Asensio, J.L.; Brown, T.; Lane, A.N. Solution Conformation of a Parallel DNA Triple Helix with 5′ and 3′ Triplex–Duplex Junctions. Structure 1999, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Besch, R.; Giovannangeli, C.; Degitz, K. Triplex-Forming Oligonucleotides—Sequence-Specific DNA Ligands as Tools for Gene Inhibition and for Modulation of DNA-Associated Functions. Curr. Drug Targets 2004, 5, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q. The Capacity to Form H-DNA Cannot Substitute for GAGA Factor Binding to a (CT)Nmiddle Dot(GA)n Regulatory Site. Nucleic Acids Res. 2003, 31, 2483–2494. [Google Scholar] [CrossRef]
- Vasquez, K.M.; Narayanan, L.; Glazer, P.M. Specific Mutations Induced by Triplex-Forming Oligonucleotides in Mice. Science 2000, 290, 530–533. [Google Scholar] [CrossRef]
- Guerrini, L.; Alvarez-Puebla, R.A. Structural Recognition of Triple-Stranded DNA by Surface-Enhanced Raman Spectroscopy. Nanomaterials 2021, 11, 326. [Google Scholar] [CrossRef]
- Hu, Y.; Cecconello, A.; Idili, A.; Ricci, F.; Willner, I. Triplex DNA Nanostructures: From Basic Properties to Applications. Angew. Chem. Int. Ed. 2017, 56, 15210–15233. [Google Scholar] [CrossRef]
- Moser, H.E.; Dervan, P.B. Sequence-Specific Cleavage of Double Helical DNA by Triple Helix Formation. Science 1987, 238, 645–650. [Google Scholar] [CrossRef]
- Datta, H.J.; Chan, P.P.; Vasquez, K.M.; Gupta, R.C.; Glazer, P.M. Triplex-Induced Recombination in Human Cell-Free Extracts. J. Biol. Chem. 2001, 276, 18018–18023. [Google Scholar] [CrossRef]
- Story, R.M.; Steitz, T.A. Structure of the recA Protein–ADP Complex. Nature 1992, 355, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Roy, U.; Greene, E.C. Demystifying the D-Loop during DNA Recombination. Nature 2020, 586, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-González, M.; Willner, I. Aptamer-Functionalized Hybrid Nanostructures for Sensing, Drug Delivery, Catalysis and Mechanical Applications. Int. J. Mol. Sci. 2021, 22, 1803. [Google Scholar] [CrossRef]
- Yang, S.; Liu, W.; Wang, R. Control of the Stepwise Assembly–Disassembly of DNA Origami Nanoclusters by pH Stimuli-Responsive DNA Triplexes. Nanoscale 2019, 11, 18026–18030. [Google Scholar] [CrossRef]
- Conde, J.; Oliva, N.; Atilano, M.; Song, H.S.; Artzi, N. Self-Assembled RNA-Triple-Helix Hydrogel Scaffold for microRNA Modulation in the Tumour Microenvironment. Nat. Mater. 2016, 15, 353–363. [Google Scholar] [CrossRef]
- Celtikci, B.; Erkmen, G.K.; Dikmen, Z.G. Regulation and Effect of Telomerase and Telomeric Length in Stem Cells. Curr. Stem Cell Res. Ther. 2021, 16, 809–823. [Google Scholar] [CrossRef]
- Shammas, M.A. Telomeres, Lifestyle, Cancer, and Aging. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 28–34. [Google Scholar] [CrossRef]
- Blasco, M.A. Telomere Length, Stem Cells and Aging. Nat. Chem. Biol. 2007, 3, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zhang, Y.; Liu, D.; Songyang, Z.; Wan, M. Telomeres—Structure, Function, and Regulation. Exp. Cell Res. 2013, 319, 133–141. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Ramaswamy, S.; Neidle, S.; Haider, S.; Parkinson, G.N. Structural Insights into the Quadruplex–Duplex 3′ Interface Formed from a Telomeric Repeat: A Potential Molecular Target. J. Am. Chem. Soc. 2016, 138, 1226–1233. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of Parallel Four-Stranded Complexes by Guanine-Rich Motifs in DNA and Its Implications for Meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Debnath, M.; Ghosh, S.; Chauhan, A.; Paul, R.; Bhattacharyya, K.; Dash, J. Preferential Targeting of I-Motifs and G-Quadruplexes by Small Molecules. Chem. Sci. 2017, 8, 7448–7456. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Cohen, G.H.; Davies, D.R. X-Ray Fiber Diffraction and Model-Building Study of Polyguanylic Acid and Polyinosinic Acid. J. Mol. Biol. 1975, 92, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative Visualization of DNA G-Quadruplex Structures in Human Cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Gehring, K.; Leroy, J.-L.; Guéron, M. A Tetrameric DNA Structure with Protonated Cytosine-Cytosine Base Pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-Motif DNA Structures Are Formed in the Nuclei of Human Cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix Formation by Guanylic Acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef]
- Kudlicki, A.S. G-Quadruplexes Involving Both Strands of Genomic DNA Are Highly Abundant and Colocalize with Functional Sites in the Human Genome. PLoS ONE 2016, 11, e0146174. [Google Scholar] [CrossRef]
- Schultze, P.; Hud, N.V.; Smith, F.W.; Feigon, J. The Effect of Sodium, Potassium and Ammonium Ions on the Conformation of the Dimeric Quadruplex Formed by the Oxytricha Nova Telomere Repeat Oligonucleotide d(G4T4G4). Nucleic Acids Res. 1999, 27, 3018–3028. [Google Scholar] [CrossRef]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and Kinetics of G-Quadruplex Structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [PubMed]
- Gavathiotis, E.; Searle, M.S. Structure of the Parallel-Stranded DNA Quadruplex d(TTAGGGT)4 Containing the Human Telomeric Repeat: Evidence for A-Tetrad Formation from NMR and Molecular Dynamics Simulations. Org. Biomol. Chem. 2003, 1, 1650–1656. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-Quadruplexes in Promoters throughout the Human Genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human Telomere, Oncogenic Promoter and 5’-UTR G-Quadruplexes: Diverse Higher Order DNA and RNA Targets for Cancer Therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef]
- Nakanishi, C.; Seimiya, H. G-Quadruplex in Cancer Biology and Drug Discovery. Biochem. Biophys. Res. Commun. 2020, 531, 45–50. [Google Scholar] [CrossRef]
- Jana, J.; Mohr, S.; Vianney, Y.M.; Weisz, K. Structural Motifs and Intramolecular Interactions in Non-Canonical G-Quadruplexes. RSC Chem. Biol. 2021, 2, 338–353. [Google Scholar] [CrossRef]
- Dumas, A.; Luedtke, N.W. Cation-Mediated Energy Transfer in G-Quadruplexes Revealed by an Internal Fluorescent Probe. J. Am. Chem. Soc. 2010, 132, 18004–18007. [Google Scholar] [CrossRef]
- Xu, Y. Formation of the G-Quadruplex and i-Motif Structures in Retinoblastoma Susceptibility Genes (Rb). Nucleic Acids Res. 2006, 34, 949–954. [Google Scholar] [CrossRef]
- Dias, E.; Battiste, J.L.; Williamson, J.R. Chemical Probe for Glycosidic Conformation in Telomeric DNAs. J. Am. Chem. Soc. 1994, 116, 4479–4480. [Google Scholar] [CrossRef]
- Risitano, A. Influence of Loop Size on the Stability of Intramolecular DNA Quadruplexes. Nucleic Acids Res. 2004, 32, 2598–2606. [Google Scholar] [CrossRef] [PubMed]
- Pendino, F.; Tarkanyi, I.; Dudognon, C.; Hillion, J.; Lanotte, M.; Aradi, J.; Segal-Bendirdjian, E. Telomeres and Telomerase: Pharmacological Targets for New Anticancer Strategies? Curr. Cancer Drug Targets 2006, 6, 147–180. [Google Scholar] [CrossRef] [PubMed]
- Hurley, L.H. DNA and Its Associated Processes as Targets for Cancer Therapy. Nat. Rev. Cancer 2002, 2, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Hirashima, K.; Seimiya, H. Telomeric Repeat-Containing RNA/G-Quadruplex-Forming Sequences Cause Genome-Wide Alteration of Gene Expression in Human Cancer Cells in Vivo. Nucleic Acids Res. 2015, 43, 2022–2032. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.-Y.; Wang, Z.-F.; Chien, C.-H.; Chang, T.-C. In-Cell Optical Imaging of Exogenous G-Quadruplex DNA by Fluorogenic Ligands. Nucleic Acids Res. 2013, 41, 10605–10618. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA Secondary Structures: Stability and Function of G-Quadruplex Structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic Acid Ligands with Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol. Ther.-Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef]
- Gatto, B.; Palumbo, M.; Sissi, C. Nucleic Acid Aptamers Based on the G-Quadruplex Structure: Therapeutic and Diagnostic Potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and Development of the G-Rich Oligonucleotide AS1411 as a Novel Treatment for Cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef]
- Yu, Y.; Liang, C.; Lv, Q.; Li, D.; Xu, X.; Liu, B.; Lu, A.; Zhang, G. Molecular Selection, Modification and Development of Therapeutic Oligonucleotide Aptamers. Int. J. Mol. Sci. 2016, 17, 358. [Google Scholar] [CrossRef]
- Soundararajan, S.; Wang, L.; Sridharan, V.; Chen, W.; Courtenay-Luck, N.; Jones, D.; Spicer, E.K.; Fernandes, D.J. Plasma Membrane Nucleolin Is a Receptor for the Anticancer Aptamer AS1411 in MV4-11 Leukemia Cells. Mol. Pharmacol. 2009, 76, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, J.; Wu, M.; Zhao, J.X. Aptamers: Active Targeting Ligands for Cancer Diagnosis and Therapy. Theranostics 2015, 5, 322–344. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Šalipur, F.R.; Shams, M.; Forsthoefel, M.K.; Bates, P.J. Mechanistic Studies of Anticancer Aptamer AS1411 Reveal a Novel Role for Nucleolin in Regulating Rac1 Activation. Mol. Oncol. 2015, 9, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A Phase II Trial of AS1411 (a Novel Nucleolin-Targeted DNA Aptamer) in Metastatic Renal Cell Carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-Quadruplex-Based Aptamers against Protein Targets in Therapy and Diagnostics. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Butovskaya, E.; Lago, S.; Garzino-Demo, A.; Pannecouque, C.; Palù, G.; Richter, S.N. The G-Quadruplex-Forming Aptamer AS1411 Potently Inhibits HIV-1 Attachment to the Host Cell. Int. J. Antimicrob. Agents 2016, 47, 311–316. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A New Paradigm for Aptamer Therapeutic AS1411 Action: Uptake by Macropinocytosis and Its Stimulation by a Nucleolin-Dependent Mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-Quadruplex-Forming Aptamers—Characteristics, Applications, and Perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef]
- Joachimi, A.; Benz, A.; Hartig, J.S. A Comparison of DNA and RNA Quadruplex Structures and Stabilities. Bioorganic Med. Chem. 2009, 17, 6811–6815. [Google Scholar] [CrossRef]
- Pagano, B.; Mattia, C.A.; Cavallo, L.; Uesugi, S.; Giancola, C.; Fraternali, F. Stability and Cations Coordination of DNA and RNA 14-Mer G-Quadruplexes: A Multiscale Computational Approach. J. Phys. Chem. B 2008, 112, 12115–12123. [Google Scholar] [CrossRef]
- Zaccaria, F.; Fonseca Guerra, C. RNA versus DNA G-Quadruplex: The Origin of Increased Stability. Chem. Eur. J. 2018, 24, 16315–16322. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N. G4-associated Human Diseases. EMBO Rep. 2015, 16, 910–922. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Xiao, S.; Liu, J.; Zhang, J.; Hao, Y.; Tan, Z. Co-Transcriptional Formation of DNA:RNA Hybrid G-Quadruplex and Potential Function as Constitutional Cis Element for Transcription Control. Nucleic Acids Res. 2013, 41, 5533–5541. [Google Scholar] [CrossRef]
- Abiri, A.; Lavigne, M.; Rezaei, M.; Nikzad, S.; Zare, P.; Mergny, J.-L.; Rahimi, H.-R. Unlocking G-Quadruplexes as Antiviral Targets. Pharmacol. Rev. 2021, 73, 897–923. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. Viral G-Quadruplexes: New Frontiers in Virus Pathogenesis and Antiviral Therapy. In Annual Reports in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 54, pp. 101–131. ISBN 978-0-12-821017-8. [Google Scholar]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More Than Just a Kink in Microbial Genomes. Trends Microbiol. 2019, 27, 148–163. [Google Scholar] [CrossRef]
- Yadav, P.; Kim, N.; Kumari, M.; Verma, S.; Sharma, T.K.; Yadav, V.; Kumar, A. G-Quadruplex Structures in Bacteria: Biological Relevance and Potential as an Antimicrobial Target. J. Bacteriol. 2021, 203, e0057720. [Google Scholar] [CrossRef]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.-L. G-Quadruplexes in Viruses: Function and Potential Therapeutic Applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Zhang, W.; Umar, M.I.; Wong, H.Y.; Seng, Z.; Xie, Y.; Zhang, Y.; Yang, L.; Kwok, C.K.; Deng, X. RNA G-Quadruplex Structures Mediate Gene Regulation in Bacteria. mBio 2020, 11, e02926-19. [Google Scholar] [CrossRef]
- Tlučková, K.; Marušič, M.; Tóthová, P.; Bauer, L.; Šket, P.; Plavec, J.; Viglasky, V. Human Papillomavirus G-Quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem. Int. Ed. 2021, 60, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A Web-Based Server for Predicting G-Quadruplexes in Nucleotide Sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Bartas, M.; Fojta, M.; Šťastný, J.; Mergny, J.-L. G4Hunter Web Application: A Web Server for G-Quadruplex Prediction. Bioinformatics 2019, 35, 3493–3495. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.K.; Abraham, J.K.; Mani, P.; Kulshrestha, R.; Chowdhury, S. QuadBase: Genome-Wide Database of G4 DNA Occurrence and Conservation in Human, Chimpanzee, Mouse and Rat Promoters and 146 Microbes. Nucleic Acids Res. 2007, 36, D381–D385. [Google Scholar] [CrossRef]
- Labudová, D.; Hon, J.; Lexa, M. Pqsfinder Web: G-Quadruplex Prediction Using Optimized Pqsfinder Algorithm. Bioinformatics 2020, 36, 2584–2586. [Google Scholar] [CrossRef] [PubMed]
- Di Antonio, M.; Rodriguez, R.; Balasubramanian, S. Experimental Approaches to Identify Cellular G-Quadruplex Structures and Functions. Methods 2012, 57, 84–92. [Google Scholar] [CrossRef]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 Helicase Resolves G-Quadruplex and Is Involved in MYC Gene Transcriptional Activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef] [PubMed]
- Konig, J.; Zarnack, K.; Rot, G.; Curk, T.; Kayikci, M.; Zupan, B.; Turner, D.J.; Luscombe, N.M.; Ule, J. iCLIP—Transcriptome-Wide Mapping of Protein-RNA Interactions with Individual Nucleotide Resolution. J. Vis. Exp. 2011, 50, 2638. [Google Scholar] [CrossRef]
- Herdy, B.; Mayer, C.; Varshney, D.; Marsico, G.; Murat, P.; Taylor, C.; D’Santos, C.; Tannahill, D.; Balasubramanian, S. Analysis of NRAS RNA G-Quadruplex Binding Proteins Reveals DDX3X as a Novel Interactor of Cellular G-Quadruplex Containing Transcripts. Nucleic Acids Res. 2018, 46, 11592–11604. [Google Scholar] [CrossRef]
- Genest, P.-A.; Baugh, L.; Taipale, A.; Zhao, W.; Jan, S.; van Luenen, H.G.A.M.; Korlach, J.; Clark, T.; Luong, K.; Boitano, M.; et al. Defining the Sequence Requirements for the Positioning of Base J in DNA Using SMRT Sequencing. Nucleic Acids Res. 2015, 43, 2102–2115. [Google Scholar] [CrossRef]
- Renaud De La Faverie, A.; Guédin, A.; Bedrat, A.; Yatsunyk, L.A.; Mergny, J.-L. Thioflavin T as a Fluorescence Light-up Probe for G4 Formation. Nucleic Acids Res. 2014, 42, e65. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Wang, L.; Liu, Y.; Chen, H.; Li, Q.; Guan, A.; Liu, M.; Tang, Y. Real-Time Monitoring of DNA G-Quadruplexes in Living Cells with a Small-Molecule Fluorescent Probe. Nucleic Acids Res. 2018, 46, 7522–7532. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Flaherty, D.P.; Chen, L.; Yang, D. High-Throughput Screening of G-Quadruplex Ligands by FRET Assay. In G-Quadruplex Nucleic Acids; Yang, D., Lin, C., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 2035, pp. 323–331. ISBN 978-1-4939-9665-0. [Google Scholar]
- Benabou, S.; Aviñó, A.; Eritja, R.; González, C.; Gargallo, R. Fundamental Aspects of the Nucleic Acid I-Motif Structures. RSC Adv. 2014, 4, 26956–26980. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, Z.; Liu, D. DNA Nanotechnology Based on I-Motif Structures. Acc. Chem. Res. 2014, 47, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Malliavin, T.E.; Gau, J.; Snoussi, K.; Leroy, J.-L. Stability of the I-Motif Structure Is Related to the Interactions between Phosphodiester Backbones. Biophys. J. 2003, 84, 3838–3847. [Google Scholar] [CrossRef]
- Yang, B.; Rodgers, M.T. Base-Pairing Energies of Proton-Bound Heterodimers of Cytosine and Modified Cytosines: Implications for the Stability of DNA i -Motif Conformations. J. Am. Chem. Soc. 2014, 136, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Bhavsar-Jog, Y.P.; Van Dornshuld, E.; Brooks, T.A.; Tschumper, G.S.; Wadkins, R.M. Epigenetic Modification, Dehydration, and Molecular Crowding Effects on the Thermodynamics of i-Motif Structure Formation from C-Rich DNA. Biochemistry 2014, 53, 1586–1594. [Google Scholar] [CrossRef]
- Lannes, L.; Halder, S.; Krishnan, Y.; Schwalbe, H. Tuning the pH Response of i-Motif DNA Oligonucleotides. ChemBioChem 2015, 16, 1647–1656. [Google Scholar] [CrossRef]
- Wright, E.P.; Abdelhamid, M.A.S.; Ehiabor, M.O.; Grigg, M.C.; Irving, K.; Smith, N.M.; Waller, Z.A.E. Epigenetic Modification of Cytosines Fine Tunes the Stability of I-Motif DNA. Nucleic Acids Res. 2020, 48, 55–62. [Google Scholar] [CrossRef]
- Abou Assi, H.; Garavís, M.; González, C.; Damha, M.J. I-Motif DNA: Structural Features and Significance to Cell Biology. Nucleic Acids Res. 2018, 46, 8038–8056. [Google Scholar] [CrossRef]
- Mergny, J. Kinetics and Thermodynamics of I-DNA Formation: Phosphodiester versus Modified Oligodeoxynucleotides. Nucleic Acids Res. 1998, 26, 4797–4803. [Google Scholar] [CrossRef] [PubMed]
- Krishnan-Ghosh, Y.; Stephens, E.; Balasubramanian, S. PNA Forms an I-Motif. Chem. Commun. 2005, 5278. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Rogers, R.A.; Zhu, J.; Zhu, J.; Burton, A.D.; Carlisle, C.B.; Burrows, C.J. 4n –1 Is a “Sweet Spot” in DNA i-Motif Folding of 2′-Deoxycytidine Homopolymers. J. Am. Chem. Soc. 2017, 139, 4682–4689. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.P.; Huppert, J.L.; Waller, Z.A.E. Identification of Multiple Genomic DNA Sequences Which Form I-Motif Structures at Neutral pH. Nucleic Acids Res. 2017, 45, 2951–2959. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Waltman, P.; Le, V.; Lewis, E. The Effect of Molecular Crowding on the Stability of Human C-MYC Promoter Sequence I-Motif at Neutral pH. Molecules 2013, 18, 12751–12767. [Google Scholar] [CrossRef]
- Miyoshi, D.; Matsumura, S.; Nakano, S.; Sugimoto, N. Duplex Dissociation of Telomere DNAs Induced by Molecular Crowding. J. Am. Chem. Soc. 2004, 126, 165–169. [Google Scholar] [CrossRef]
- Alba, J.J.; Sadurní, A.; Gargallo, R. Nucleic Acid i- Motif Structures in Analytical Chemistry. Crit. Rev. Anal. Chem. 2016, 46, 443–454. [Google Scholar] [CrossRef]
- Liu, D.; Balasubramanian, S. A Proton-Fuelled DNA Nanomachine. Angew. Chem. Int. Ed. 2003, 42, 5734–5736. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-G.; Elbaz, J.; Willner, I. DNA Machines: Bipedal Walker and Stepper. Nano Lett. 2011, 11, 304–309. [Google Scholar] [CrossRef]
- Zhao, D.; Tang, H.; Wang, H.; Yang, C.; Li, Y. Analytes Triggered Conformational Switch of I-Motif DNA inside Gold-Decorated Solid-State Nanopores. ACS Sens. 2020, 5, 2177–2183. [Google Scholar] [CrossRef]
- Li, T.; Ackermann, D.; Hall, A.M.; Famulok, M. Input-Dependent Induction of Oligonucleotide Structural Motifs for Performing Molecular Logic. J. Am. Chem. Soc. 2012, 134, 3508–3516. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, G.; Liu, H.; Li, D.; Fan, C.; Liu, D. An Electrochemically Actuated Reversible DNA Switch. Nano Lett. 2010, 10, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; MG, S.; Goswami, D.; Gupta, G.D.; Mayor, S.; Krishnan, Y. A DNA Nanomachine That Maps Spatial and Temporal pH Changes inside Living Cells. Nat. Nanotechnol. 2009, 4, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.A.; Chimenti, M.; Jacobson, M.P.; Barber, D.L. Dysregulated pH: A Perfect Storm for Cancer Progression. Nat. Rev. Cancer 2011, 11, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wei, C.; Jia, G.; Wang, X.; Feng, Z.; Li, C. Formation of I-Motif Structure at Neutral and Slightly Alkaline pH. Mol. BioSyst. 2010, 6, 580–586. [Google Scholar] [CrossRef]
- Kim, B.G.; Chalikian, T.V. Thermodynamic Linkage Analysis of pH-Induced Folding and Unfolding Transitions of i-Motifs. Biophys. Chem. 2016, 216, 19–22. [Google Scholar] [CrossRef]
- Rajendran, A.; Nakano, S.; Sugimoto, N. Molecular Crowding of the Cosolutes Induces an Intramolecular I-Motif Structure of Triplet Repeat DNA Oligomers at Neutral pH. Chem. Commun. 2010, 46, 1299. [Google Scholar] [CrossRef]
- Sun, D.; Hurley, L.H. The Importance of Negative Superhelicity in Inducing the Formation of G-Quadruplex and i-Motif Structures in the c-Myc Promoter: Implications for Drug Targeting and Control of Gene Expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.; Fábián, L.; MacDonald, C.J.; Cheesman, M.R.; Gates, A.J.; Waller, Z.A. Redox-Dependent Control of i-Motif DNA Structure Using Copper Cations. Nucleic Acids Res. 2018, 46, 5886–5893. [Google Scholar] [CrossRef]
- Day, H.A.; Huguin, C.; Waller, Z.A.E. Silver Cations Fold I-Motif at Neutral pH. Chem. Commun. 2013, 49, 7696. [Google Scholar] [CrossRef]
- Satpathi, S.; Sappati, S.; Das, K.; Hazra, P. Structural Characteristics Requisite for the Ligand-Based Selective Detection of i-Motif DNA. Org. Biomol. Chem. 2019, 17, 5392–5399. [Google Scholar] [CrossRef] [PubMed]
- Wolski, P.; Nieszporek, K.; Panczyk, T. Regulation of Water Access, Storage, Separation and Release of Drugs from the Carbon Nanotube Functionalized by Cytosine Rich DNA Fragments. Biomater. Adv. 2022, 137, 212835. [Google Scholar] [CrossRef] [PubMed]
- Wolski, P.; Nieszporek, K.; Panczyk, T. Carbon Nanotubes and Short Cytosine-Rich Telomeric DNA Oligomeres as Platforms for Controlled Release of Doxorubicin—A Molecular Dynamics Study. Int. J. Mol. Sci. 2020, 21, 3619. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-Y.; Vankayalapati, H.; Shin-ya, K.; Wierzba, K.; Hurley, L.H. Telomestatin, a Potent Telomerase Inhibitor That Interacts Quite Specifically with the Human Telomeric Intramolecular G-Quadruplex. J. Am. Chem. Soc. 2002, 124, 2098–2099. [Google Scholar] [CrossRef]
- Shin-ya, K.; Wierzba, K.; Matsuo, K.; Ohtani, T.; Yamada, Y.; Furihata, K.; Hayakawa, Y.; Seto, H. Telomestatin, a Novel Telomerase Inhibitor from Streptomyces Anulatus. J. Am. Chem. Soc. 2001, 123, 1262–1263. [Google Scholar] [CrossRef]
- Anantha, N.V.; Azam, M.; Sheardy, R.D. Porphyrin Binding to Quadruplexed T4G4. Biochemistry 1998, 37, 2709–2714. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-Quadruplex-Interactive Molecule BRACO-19 Inhibits Tumor Growth, Consistent with Telomere Targeting and Interference with Telomerase Function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef]
- Rodriguez, R.; Müller, S.; Yeoman, J.A.; Trentesaux, C.; Riou, J.-F.; Balasubramanian, S. A Novel Small Molecule That Alters Shelterin Integrity and Triggers a DNA-Damage Response at Telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759. [Google Scholar] [CrossRef]
- De Cian, A.; DeLemos, E.; Mergny, J.-L.; Teulade-Fichou, M.-P.; Monchaud, D. Highly Efficient G-Quadruplex Recognition by Bisquinolinium Compounds. J. Am. Chem. Soc. 2007, 129, 1856–1857. [Google Scholar] [CrossRef]
- Tera, M.; Iida, K.; Ishizuka, H.; Takagi, M.; Suganuma, M.; Doi, T.; Shin-ya, K.; Nagasawa, K. Synthesis of a Potent G-Quadruplex-Binding Macrocyclic Heptaoxazole. ChemBioChem 2009, 10, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Sasaki, I.; Tsukakoshi, K.; Ma, Y.; Nagasawa, K.; Numata, S.; Inoue, Y.; Kim, Y.; Ikebukuro, K. Detection of CpG Methylation in G-Quadruplex Forming Sequences Using G-Quadruplex Ligands. Int. J. Mol. Sci. 2021, 22, 13159. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-Quadruplexes in Gene Promoters: A Novel Anticancer Strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, S.; Gu, Y.; Yan, Y.; Stovall, D.B.; Li, D.; Sui, G. Human MYC G-Quadruplex: From Discovery to a Cancer Therapeutic Target. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1874, 188410. [Google Scholar] [CrossRef]
- Dutta, D.; Debnath, M.; Müller, D.; Paul, R.; Das, T.; Bessi, I.; Schwalbe, H.; Dash, J. Cell Penetrating Thiazole Peptides Inhibit C-MYC Expression via Site-Specific Targeting of c-MYC G-Quadruplex. Nucleic Acids Res. 2018, 46, 5355–5365. [Google Scholar] [CrossRef]
- Palumbo, S.L.; Ebbinghaus, S.W.; Hurley, L.H. Formation of a Unique End-to-End Stacked Pair of G-Quadruplexes in the hTERT Core Promoter with Implications for Inhibition of Telomerase by G-Quadruplex-Interactive Ligands. J. Am. Chem. Soc. 2009, 131, 10878–10891. [Google Scholar] [CrossRef] [PubMed]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA Quadruplex Formation within the Human c-Kit Oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef]
- Morgan, R.K.; Batra, H.; Gaerig, V.C.; Hockings, J.; Brooks, T.A. Identification and Characterization of a New G-Quadruplex Forming Region within the kRAS Promoter as a Transcriptional Regulator. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2016, 1859, 235–245. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the Structural and Drug-Recognition Complexity of the G-Quadruplex-Forming Region Upstream of the Bcl-2 P1 Promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef]
- Tian, T.; Chen, Y.-Q.; Wang, S.-R.; Zhou, X. G-Quadruplex: A Regulator of Gene Expression and Its Chemical Targeting. Chem 2018, 4, 1314–1344. [Google Scholar] [CrossRef]
- Scott, L.; Chalikian, T.V. Stabilization of G-Quadruplex-Duplex Hybrid Structures Induced by Minor Groove-Binding Drugs. Life 2022, 12, 597. [Google Scholar] [CrossRef] [PubMed]
- Libera, V.; Ripanti, F.; Petrillo, C.; Sacchetti, F.; Ramos-Soriano, J.; Galan, M.C.; Schirò, G.; Paciaroni, A.; Comez, L. Stability of Human Telomeric G-Quadruplexes Complexed with Photosensitive Ligands and Irradiated with Visible Light. Int. J. Mol. Sci. 2023, 24, 9090. [Google Scholar] [CrossRef] [PubMed]
- Pandya, N.; Rani, R.; Kumar, V.; Kumar, A. Discovery of a Potent Guanidine Derivative That Selectively Binds and Stabilizes the Human BCL-2 G-Quadruplex DNA and Downregulates the Transcription. Gene 2023, 851, 146975. [Google Scholar] [CrossRef] [PubMed]
- Mazzini, S.; Princiotto, S.; Artali, R.; Musso, L.; Aviñó, A.; Eritja, R.; Gargallo, R.; Dallavalle, S. Exploring the Interaction of G-Quadruplex Binders with a (3 + 1) Hybrid G-Quadruplex Forming Sequence within the PARP1 Gene Promoter Region. Molecules 2022, 27, 4792. [Google Scholar] [CrossRef] [PubMed]
- Nocentini, A.; Di Porzio, A.; Bonardi, A.; Bazzicalupi, C.; Petreni, A.; Biver, T.; Bua, S.; Marzano, S.; Amato, J.; Pagano, B.; et al. Development of a Multi-Targeted Chemotherapeutic Approach Based on G-Quadruplex Stabilisation and Carbonic Anhydrase Inhibition. J. Enzym. Inhib. Med. Chem. 2024, 39, 2366236. [Google Scholar] [CrossRef]
- Pont, I.; Galiana-Roselló, C.; Sabater-Arcis, M.; Bargiela, A.; Frías, J.C.; Albelda, M.T.; González-García, J.; García-España, E. Development of Potent Tripodal G-Quadruplex DNA Binders and Their Efficient Delivery to Cancer Cells by Aptamer Functionalised Liposomes. Org. Biomol. Chem. 2023, 21, 1000–1007. [Google Scholar] [CrossRef]
- Manna, S.; Srivatsan, S.G. Fluorescence-Based Tools to Probe G-Quadruplexes in Cell-Free and Cellular Environments. RSC Adv. 2018, 8, 25673–25694. [Google Scholar] [CrossRef]
- Figueiredo, J.; Santos, T.; Miranda, A.; Alexandre, D.; Teixeira, B.; Simões, P.; Lopes-Nunes, J.; Cruz, C. Ligands as Stabilizers of G-Quadruplexes in Non-Coding RNAs. Molecules 2021, 26, 6164. [Google Scholar] [CrossRef]
- Renard, I.; Grandmougin, M.; Roux, A.; Yang, S.Y.; Lejault, P.; Pirrotta, M.; Wong, J.M.Y.; Monchaud, D. Small-Molecule Affinity Capture of DNA/RNA Quadruplexes and Their Identification in Vitro and in Vivo through the G4RP Protocol. Nucleic Acids Res. 2019, 47, 5502–5510. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Y.; Zhang, W. G-Quadruplex Structures and Their Interaction Diversity with Ligands. ChemMedChem 2014, 9, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Lejault, P.; Prudent, L.; Terrier, M.-P.; Perreault, J.-P. Small Molecule Chaperones Facilitate the Folding of RNA G-Quadruplexes. Biochimie 2023, 214, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, A.; Hukezalie, K.; Winckler, P.; Katranji, F.; Chanteloup, G.; Pirrotta, M.; Perrier-Cornet, J.-M.; Wong, J.M.Y.; Monchaud, D. Visualization of RNA-Quadruplexes in Live Cells. J. Am. Chem. Soc. 2015, 137, 8521–8525. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, O.Y.; Rangan, A.; Chemeris, V.V.; Hurley, L.H. Cationic Porphyrins Promote the Formation of I-Motif DNA and Bind Peripherally by a Nonintercalative Mechanism. Biochemistry 2000, 39, 15083–15090. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, Y.; Chen, T.; Wei, C. The Interactions of Phenanthroline Compounds with DNAs: Preferential Binding to Telomeric Quadruplex over Duplex. Int. J. Biol. Macromol. 2013, 52, 1–8. [Google Scholar] [CrossRef]
- Xue, L.; Ranjan, N.; Arya, D.P. Synthesis and Spectroscopic Studies of the Aminoglycoside (Neomycin)−Perylene Conjugate Binding to Human Telomeric DNA. Biochemistry 2011, 50, 2838–2849. [Google Scholar] [CrossRef]
- Ma, D.-L.; Kwan, M.H.-T.; Chan, D.S.-H.; Lee, P.; Yang, H.; Ma, V.P.-Y.; Bai, L.-P.; Jiang, Z.-H.; Leung, C.-H. Crystal Violet as a Fluorescent Switch-on Probe for i-Motif: Label-Free DNA-Based Logic Gate. Analyst 2011, 136, 2692. [Google Scholar] [CrossRef]
- Gao, R.-R.; Shi, S.; Zhu, Y.; Huang, H.-L.; Yao, T.-M. A RET-Supported Logic Gate Combinatorial Library to Enable Modeling and Implementation of Intelligent Logic Functions. Chem. Sci. 2016, 7, 1853–1861. [Google Scholar] [CrossRef]
- Xu, L.; Hong, S.; Sun, N.; Wang, K.; Zhou, L.; Ji, L.; Pei, R. Berberine as a Novel Light-up i-Motif Fluorescence Ligand and Its Application in Designing Molecular Logic Systems. Chem. Commun. 2016, 52, 179–182. [Google Scholar] [CrossRef]
- Kendrick, S.; Kang, H.-J.; Alam, M.P.; Madathil, M.M.; Agrawal, P.; Gokhale, V.; Yang, D.; Hecht, S.M.; Hurley, L.H. The Dynamic Character of the BCL2 Promoter i-Motif Provides a Mechanism for Modulation of Gene Expression by Compounds That Bind Selectively to the Alternative DNA Hairpin Structure. J. Am. Chem. Soc. 2014, 136, 4161–4171. [Google Scholar] [CrossRef]
- Wright, E.P.; Day, H.A.; Ibrahim, A.M.; Kumar, J.; Boswell, L.J.E.; Huguin, C.; Stevenson, C.E.M.; Pors, K.; Waller, Z.A.E. Mitoxantrone and Analogues Bind and Stabilize I-Motif Forming DNA Sequences. Sci. Rep. 2016, 6, 39456. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.V.; Wang, T.; Chappeta, V.R.; Wu, G.; Onel, B.; Chawla, R.; Quijada, H.; Camp, S.M.; Chiang, E.T.; Lassiter, Q.R.; et al. The Consequences of Overlapping G-Quadruplexes and i-Motifs in the Platelet-Derived Growth Factor Receptor β Core Promoter Nuclease Hypersensitive Element Can Explain the Unexpected Effects of Mutations and Provide Opportunities for Selective Targeting of Both Structures by Small Molecules to Downregulate Gene Expression. J. Am. Chem. Soc. 2017, 139, 7456–7475. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Cao, J.; Kuang, G.; Qiu, J.; Zhang, M.; Zhang, Y.; Wang, M.; Li, X.; Kang, S.; Ou, T.-M.; et al. Syntheses and Evaluation of New Acridone Derivatives for Selective Binding of Oncogene c-Myc Promoter i-Motifs in Gene Transcriptional Regulation. Chem. Commun. 2018, 54, 2036–2039. [Google Scholar] [CrossRef] [PubMed]
- Dzatko, S.; Krafcikova, M.; Hänsel-Hertsch, R.; Fessl, T.; Fiala, R.; Loja, T.; Krafcik, D.; Mergny, J.; Foldynova-Trantirkova, S.; Trantirek, L. Evaluation of the Stability of DNA i-Motifs in the Nuclei of Living Mammalian Cells. Angew. Chem. Int. Ed. 2018, 57, 2165–2169. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qu, K.; Zhao, C.; Wu, L.; Ren, J.; Wang, J.; Qu, X. Insights into the Biomedical Effects of Carboxylated Single-Wall Carbon Nanotubes on Telomerase and Telomeres. Nat. Commun. 2012, 3, 1074. [Google Scholar] [CrossRef]
- Li, X.; Peng, Y.; Ren, J.; Qu, X. Carboxyl-Modified Single-Walled Carbon Nanotubes Selectively Induce Human Telomeric i-Motif Formation. Proc. Natl. Acad. Sci. USA 2006, 103, 19658–19663. [Google Scholar] [CrossRef]
- Sun, H.; Ren, J.; Qu, X. Carbon Nanomaterials and DNA: From Molecular Recognition to Applications. Acc. Chem. Res. 2016, 49, 461–470. [Google Scholar] [CrossRef]
- Zhao, C.; Ren, J.; Qu, X. Single-Walled Carbon Nanotubes Binding to Human Telomeric i-Motif DNA Under Molecular-Crowding Conditions: More Water Molecules Released. Chem. A Eur. J. 2008, 14, 5435–5439. [Google Scholar] [CrossRef]
- Saha, P.; Panda, D.; Müller, D.; Maity, A.; Schwalbe, H.; Dash, J. In Situ Formation of Transcriptional Modulators Using Non-Canonical DNA i-Motifs. Chem. Sci. 2020, 11, 2058–2067. [Google Scholar] [CrossRef]
- Brown, S.L.; Kendrick, S. The I-Motif as a Molecular Target: More Than a Complementary DNA Secondary Structure. Pharmaceuticals 2021, 14, 96. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Yang, Q.-F.; Lin, X.; Chen, D.; Wang, Z.-Y.; Chen, B.; Han, H.-Y.; Chen, H.-D.; Cai, K.-C.; Li, Q.; et al. G4LDB 2.2: A Database for Discovering and Studying G-Quadruplex and i-Motif Ligands. Nucleic Acids Res. 2022, 50, D150–D160. [Google Scholar] [CrossRef] [PubMed]
- Lech, C.J.; Heddi, B.; Phan, A.T. Guanine Base Stacking in G-Quadruplex Nucleic Acids. Nucleic Acids Res. 2013, 41, 2034–2046. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.I.V.; Almodôvar, V.A.S.; Candeias, N.R.; Santos, T.; Cruz, C.; Neves, M.G.P.M.S.; Tomé, A.C. Diketopyrrolo[3,4-c]Pyrrole Derivative as a Promising Ligand for the Stabilization of G-Quadruplex DNA Structures. Bioorg. Chem. 2022, 122, 105703. [Google Scholar] [CrossRef] [PubMed]
- Clay, E.H.; Gould, I.R. A Combined QM and MM Investigation into Guanine Quadruplexes. J. Mol. Graph. Model. 2005, 24, 138–146. [Google Scholar] [CrossRef]
- Šponer, J.; Banáš, P.; Jurečka, P.; Zgarbová, M.; Kührová, P.; Havrila, M.; Krepl, M.; Stadlbauer, P.; Otyepka, M. Molecular Dynamics Simulations of Nucleic Acids. From Tetranucleotides to the Ribosome. J. Phys. Chem. Lett. 2014, 5, 1771–1782. [Google Scholar] [CrossRef]
- Salsbury, A.M.; Dean, T.J.; Lemkul, J.A. Polarizable Molecular Dynamics Simulations of Two c-Kit Oncogene Promoter G-Quadruplexes: Effect of Primary and Secondary Structure on Loop and Ion Sampling. J. Chem. Theory Comput. 2020, 16, 3430–3444. [Google Scholar] [CrossRef]
- Hazel, P.; Huppert, J.; Balasubramanian, S.; Neidle, S. Loop-Length-Dependent Folding of G-Quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. [Google Scholar] [CrossRef]
- Šponer, J.; Špačková, N. Molecular Dynamics Simulations and Their Application to Four-Stranded DNA. Methods 2007, 43, 278–290. [Google Scholar] [CrossRef]
- Chowdhury, S.; Bansal, M. G-Quadruplex Structure Can Be Stable with Only Some Coordination Sites Being Occupied by Cations: A Six-Nanosecond Molecular Dynamics Study. J. Phys. Chem. B 2001, 105, 7572–7578. [Google Scholar] [CrossRef]
- Agrawal, S.; Ojha, R.P.; Maiti, S. Energetics of the Human Tel-22 Quadruplex−Telomestatin Interaction: A Molecular Dynamics Study. J. Phys. Chem. B 2008, 112, 6828–6836. [Google Scholar] [CrossRef]
- Aggrawal, M.; Joo, H.; Liu, W.; Tsai, J.; Xue, L. 8-Oxo-7,8-Dihydrodeoxyadenosine: The First Example of a Native DNA Lesion That Stabilizes Human Telomeric G-Quadruplex DNA. Biochem. Biophys. Res. Commun. 2012, 421, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Panczyk, T.; Wojton, P.; Wolski, P. Molecular Dynamics Study of the Interaction of Carbon Nanotubes with Telomeric DNA Fragment Containing Noncanonical G-Quadruplex and i-Motif Forms. Int. J. Mol. Sci. 2020, 21, 1925. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, D.A.; Nesbitt, D.J. Kinetic and Thermodynamic Control of G-Quadruplex Polymorphism by Na + and K + Cations. J. Phys. Chem. B 2023, 127, 6842–6855. [Google Scholar] [CrossRef] [PubMed]
- Šponer, J.; Bussi, G.; Stadlbauer, P.; Kührová, P.; Banáš, P.; Islam, B.; Haider, S.; Neidle, S.; Otyepka, M. Folding of Guanine Quadruplex Molecules–Funnel-like Mechanism or Kinetic Partitioning? An Overview from MD Simulation Studies. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1246–1263. [Google Scholar] [CrossRef]
- Stadlbauer, P.; Kührová, P.; Banáš, P.; Koča, J.; Bussi, G.; Trantírek, L.; Otyepka, M.; Šponer, J. Hairpins Participating in Folding of Human Telomeric Sequence Quadruplexes Studied by Standard and T-REMD Simulations. Nucleic Acids Res. 2015, 43, gkv994. [Google Scholar] [CrossRef]
- Kim, H.; Kim, E.; Pak, Y. Computational Probing of the Folding Mechanism of Human Telomeric G-Quadruplex DNA. J. Chem. Inf. Model. 2023, 63, 6366–6375. [Google Scholar] [CrossRef]
- Bian, Y.; Song, F.; Zhang, J.; Yu, J.; Wang, J.; Wang, W. Insights into the Kinetic Partitioning Folding Dynamics of the Human Telomeric G-Quadruplex from Molecular Simulations and Machine Learning. J. Chem. Theory Comput. 2020, 16, 5936–5947. [Google Scholar] [CrossRef]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the Human Telomere in K + Solution: A Stable Basket-Type G-Quadruplex with Only Two G-Tetrad Layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar] [CrossRef]
- Husby, J.; Todd, A.K.; Platts, J.A.; Neidle, S. Small-molecule G-quadruplex Interactions: Systematic Exploration of Conformational Space Using Multiple Molecular Dynamics. Biopolymers 2013, 99, 989–1005. [Google Scholar] [CrossRef]
- Islam, B.; Sgobba, M.; Laughton, C.; Orozco, M.; Sponer, J.; Neidle, S.; Haider, S. Conformational Dynamics of the Human Propeller Telomeric DNA Quadruplex on a Microsecond Time Scale. Nucleic Acids Res. 2013, 41, 2723–2735. [Google Scholar] [CrossRef]
- Islam, B.; Stadlbauer, P.; Gil-Ley, A.; Pérez-Hernández, G.; Haider, S.; Neidle, S.; Bussi, G.; Banas, P.; Otyepka, M.; Sponer, J. Exploring the Dynamics of Propeller Loops in Human Telomeric DNA Quadruplexes Using Atomistic Simulations. J. Chem. Theory Comput. 2017, 13, 2458–2480. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.; Kleist, C.; Czub, J. Molecular Dynamics Simulations Reveal the Balance of Forces Governing the Formation of a Guanine Tetrad—A Common Structural Unit of G-Quadruplex DNA. Nucleic Acids Res. 2016, 44, 3020–3030. [Google Scholar] [CrossRef] [PubMed]
- Shweta, H.; Sen, S. Dynamics of Water and Ions around DNA: What Is so Special about Them? J. Biosci. 2018, 43, 499–518. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Gonzalez, D.; Perez-Machado, G.; Pallardo, F.; Garrigues-Pelufo, T.-M.; Cabrera-Perez, M.-A. Computational Tools in the Discovery of New G-Quadruplex Ligands with Potential Anticancer Activity. Curr. Top. Med Chem. 2013, 12, 2843–2856. [Google Scholar] [CrossRef] [PubMed]
- Monsen, R.C.; Trent, J.O. G-Quadruplex Virtual Drug Screening: A Review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef]
- Mulliri, S.; Laaksonen, A.; Spanu, P.; Farris, R.; Farci, M.; Mingoia, F.; Roviello, G.N.; Mocci, F. Spectroscopic and In Silico Studies on the Interaction of Substituted Pyrazolo[1,2-a]Benzo[1,2,3,4]Tetrazine-3-One Derivatives with c-Myc G4-DNA. Int. J. Mol. Sci. 2021, 22, 6028. [Google Scholar] [CrossRef]
- Zareie, A.R.; Dabral, P.; Verma, S.C. G-Quadruplexes in the Regulation of Viral Gene Expressions and Their Impacts on Controlling Infection. Pathogens 2024, 13, 60. [Google Scholar] [CrossRef]
- Moraca, F.; Amato, J.; Ortuso, F.; Artese, A.; Pagano, B.; Novellino, E.; Alcaro, S.; Parrinello, M.; Limongelli, V. Ligand Binding to Telomeric G-Quadruplex DNA Investigated by Funnel-Metadynamics Simulations. Proc. Natl. Acad. Sci. USA 2017, 114, E2136–E2145. [Google Scholar] [CrossRef]
- Smiatek, J.; Chen, C.; Liu, D.; Heuer, A. Stable Conformations of a Single Stranded Deprotonated DNA I-Motif. J. Phys. Chem. B 2011, 115, 13788–13795. [Google Scholar] [CrossRef]
- Singh, R.P.; Blossey, R.; Cleri, F. Structure and Mechanical Characterization of DNA I-Motif Nanowires by Molecular Dynamics Simulation. Biophys. J. 2013, 105, 2820–2831. [Google Scholar] [CrossRef]
- Smiatek, J.; Heuer, A. Deprotonation Mechanism of a Single-Stranded DNA i-Motif. RSC Adv. 2014, 4, 17110–17113. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Nakano, M.; Pramanik, S.; Tanaka, S.; Sugimoto, N. I-Motifs Are More Stable than G-Quadruplexes in a Hydrated Ionic Liquid. Chem. Commun. 2015, 51, 6909–6912. [Google Scholar] [CrossRef] [PubMed]
- Garabedian, A.; Butcher, D.; Lippens, J.L.; Miksovska, J.; Chapagain, P.P.; Fabris, D.; Ridgeway, M.E.; Park, M.A.; Fernandez-Lima, F. Structures of the Kinetically Trapped I-Motif DNA Intermediates. Phys. Chem. Chem. Phys. 2016, 18, 26691–26702. [Google Scholar] [CrossRef] [PubMed]
- Protopopova, A.D.; Tsvetkov, V.B.; Varizhuk, A.M.; Barinov, N.A.; Podgorsky, V.V.; Klinov, D.V.; Pozmogova, G.E. The Structural Diversity of C-Rich DNA Aggregates: Unusual Self-Assembly of Beetle-like Nanostructures. Phys. Chem. Chem. Phys. 2018, 20, 3543–3553. [Google Scholar] [CrossRef] [PubMed]
- Panczyk, T.; Wolski, P. Molecular Dynamics Analysis of Stabilities of the Telomeric Watson-Crick Duplex and the Associated i-Motif as a Function of pH and Temperature. Biophys. Chem. 2018, 237, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Mondal, M.; Bhattacharyya, D.; Gao, Y.Q. Structural Properties and Influence of Solvent on the Stability of Telomeric Four-Stranded i-Motif DNA. Phys. Chem. Chem. Phys. 2019, 21, 21549–21560. [Google Scholar] [CrossRef]
- Wolski, P.; Nieszporek, K.; Panczyk, T. G-Quadruplex and I-Motif Structures within the Telomeric DNA Duplex. A Molecular Dynamics Analysis of Protonation States as Factors Affecting Their Stability. J. Phys. Chem. B 2019, 123, 468–479. [Google Scholar] [CrossRef]
- Wolski, P.; Nieszporek, K.; Panczyk, T. Cytosine-Rich DNA Fragments Covalently Bound to Carbon Nanotube as Factors Triggering Doxorubicin Release at Acidic pH. A Molecular Dynamics Study. Int. J. Mol. Sci. 2021, 22, 8466. [Google Scholar] [CrossRef]
- Shamim, A.; Razzaq, M.; Kim, K.K. MD-TSPC4: Computational Method for Predicting the Thermal Stability of I-Motif. Int. J. Mol. Sci. 2020, 22, 61. [Google Scholar] [CrossRef]
- Xi, D.; Cui, M.; Zhou, X.; Zhuge, X.; Ge, Y.; Wang, Y.; Zhang, S. Nanopore-Based Single-Molecule Investigation of DNA Sequences with Potential to Form i-Motif Structures. ACS Sens. 2021, 6, 2691–2699. [Google Scholar] [CrossRef]
- Lopez, A.; Zhao, Y.; Huang, Z.; Guo, Y.; Guan, S.; Jia, Y.; Liu, J. Poly-Cytosine Deoxyribonucleic Acid Strongly Anchoring on Graphene Oxide Due to Flexible Backbone Phosphate Interactions. Adv. Mater. Interfaces 2021, 8, 2001798. [Google Scholar] [CrossRef]
- Wojton, P.; Wolski, P.; Wolinski, K.; Panczyk, T. Protonation of Cytosine-Rich Telomeric DNA Fragments by Carboxylated Carbon Nanotubes: Insights from Computational Studies. J. Phys. Chem. B 2021, 125, 5526–5536. [Google Scholar] [CrossRef] [PubMed]
- Panczyk, T.; Nieszporek, K.; Wolski, P. Stability and Existence of Noncanonical I-Motif DNA Structures in Computer Simulations Based on Atomistic and Coarse-Grained Force Fields. Molecules 2022, 27, 4915. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obara, P.; Wolski, P.; Pańczyk, T. Insights into the Molecular Structure, Stability, and Biological Significance of Non-Canonical DNA Forms, with a Focus on G-Quadruplexes and i-Motifs. Molecules 2024, 29, 4683. https://doi.org/10.3390/molecules29194683
Obara P, Wolski P, Pańczyk T. Insights into the Molecular Structure, Stability, and Biological Significance of Non-Canonical DNA Forms, with a Focus on G-Quadruplexes and i-Motifs. Molecules. 2024; 29(19):4683. https://doi.org/10.3390/molecules29194683
Chicago/Turabian StyleObara, Patrycja, Paweł Wolski, and Tomasz Pańczyk. 2024. "Insights into the Molecular Structure, Stability, and Biological Significance of Non-Canonical DNA Forms, with a Focus on G-Quadruplexes and i-Motifs" Molecules 29, no. 19: 4683. https://doi.org/10.3390/molecules29194683
APA StyleObara, P., Wolski, P., & Pańczyk, T. (2024). Insights into the Molecular Structure, Stability, and Biological Significance of Non-Canonical DNA Forms, with a Focus on G-Quadruplexes and i-Motifs. Molecules, 29(19), 4683. https://doi.org/10.3390/molecules29194683