Fluorescence Polarization Assay for Infection Diagnostics: A Review
Abstract
:1. Introduction
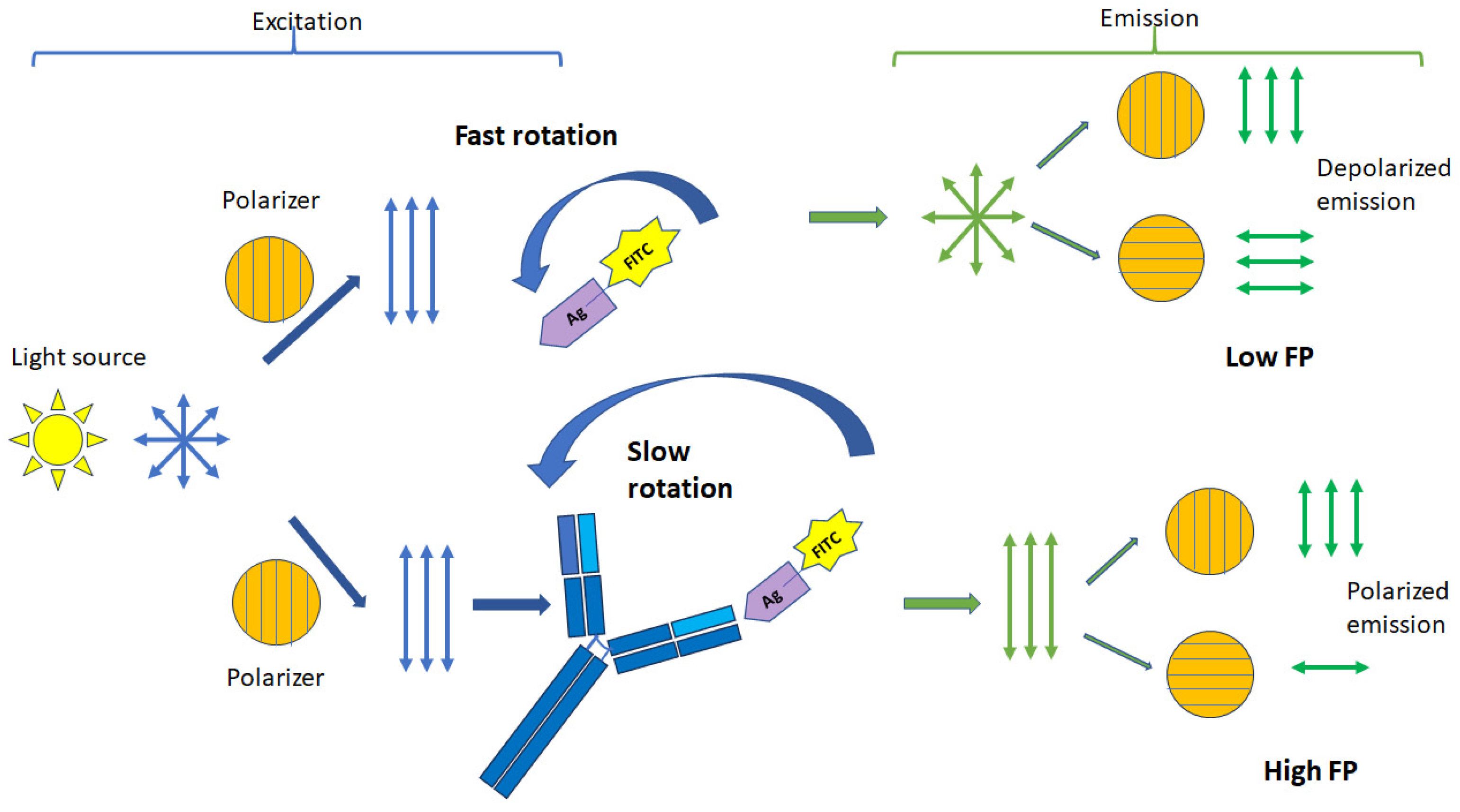
2. Principles of Fluorescence Polarization Assay
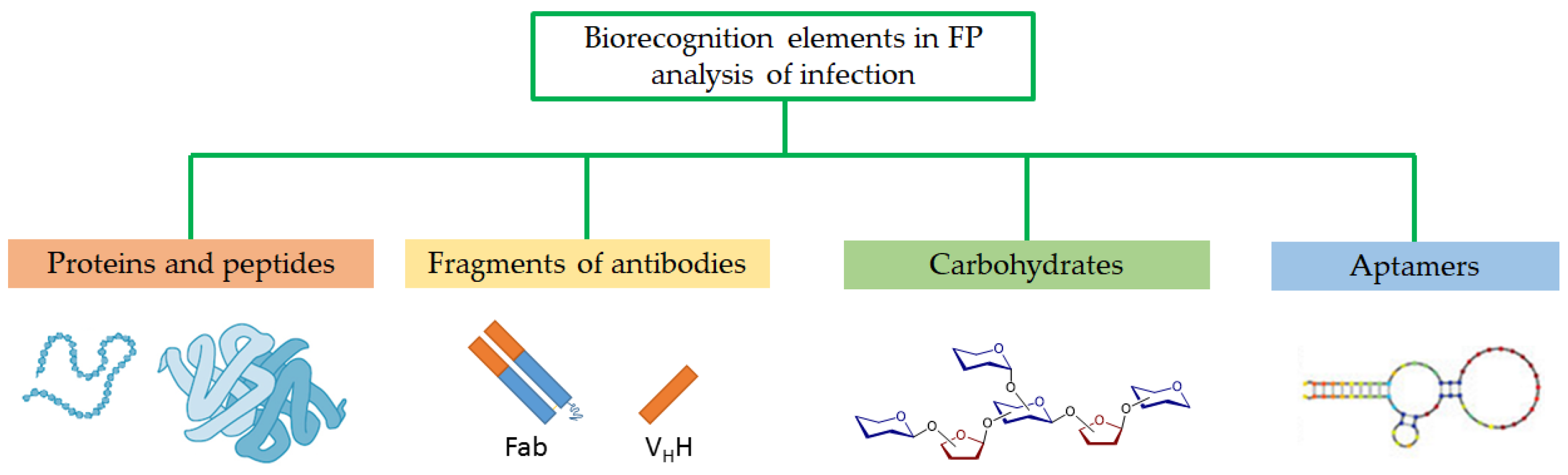
3. Detection of Infectious Diseases by FPA
4. Detection of Antibodies to Viruses and Bacteria by FPA
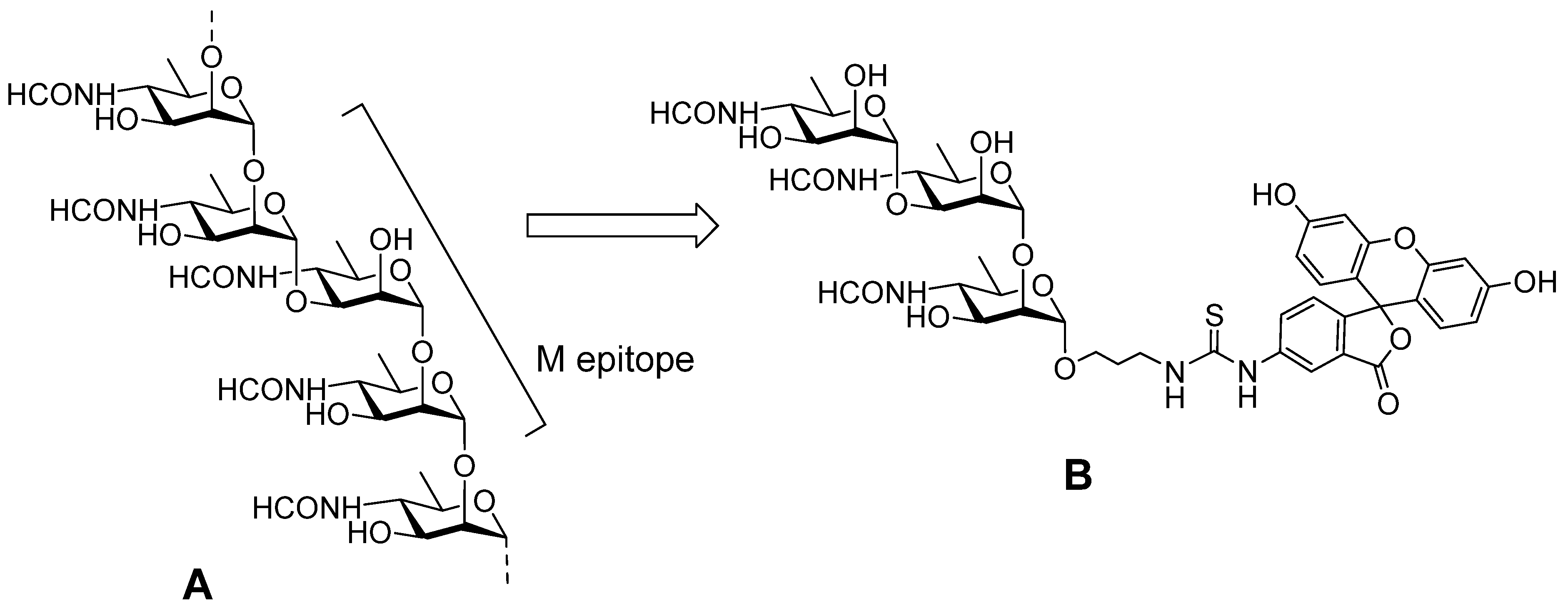
4.1. Antibodies to Brucellosis
4.2. Antibodies to Salmonella
4.3. Antibodies to Tuberculosis
4.4. Antibodies to Equine Infectious Anemia Virus (EIAV)
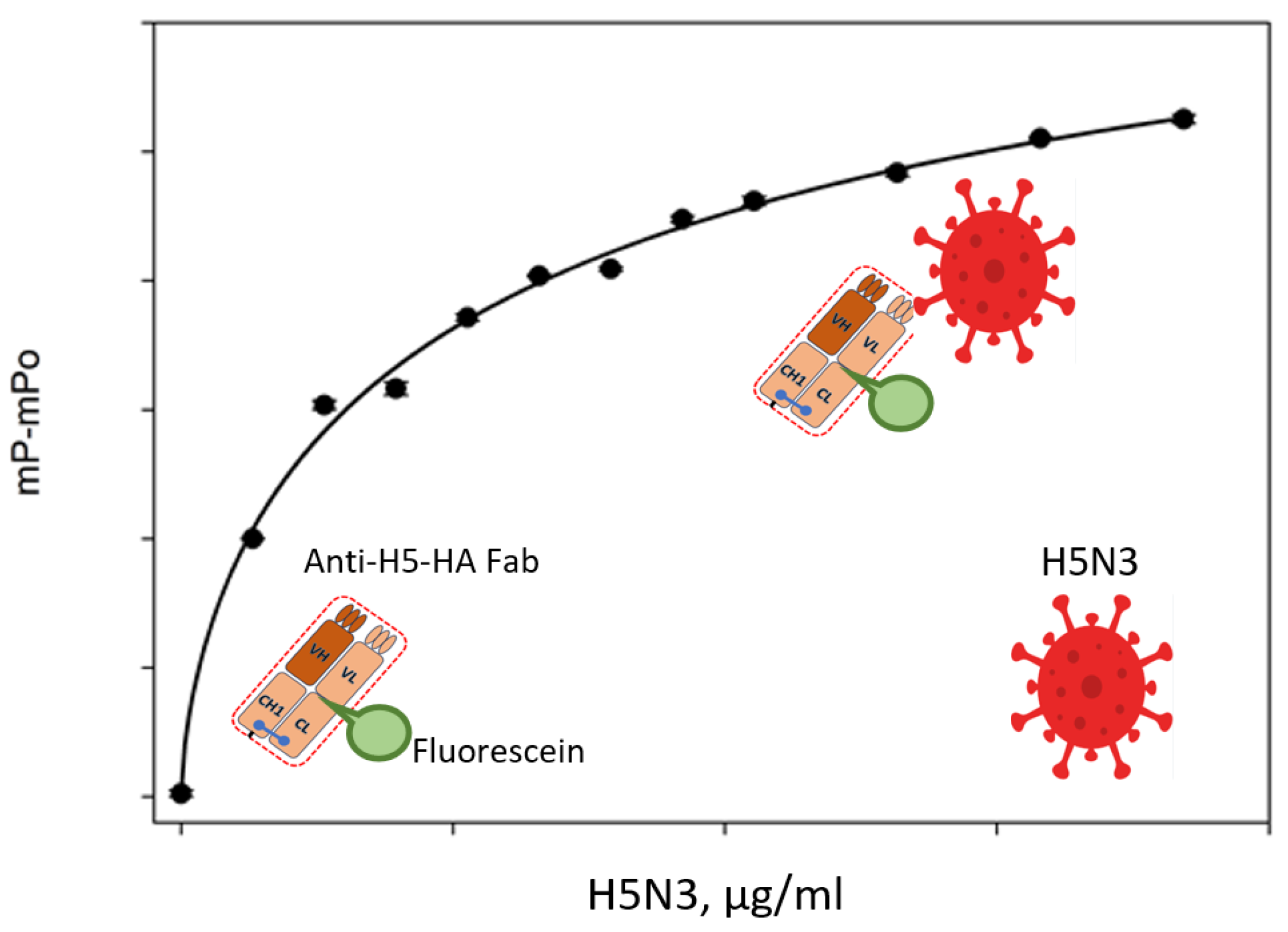
4.5. Antibodies to Avian Influenza a Virus (IAV)
4.6. Antibodies to SARS-CoV-2
| Disease | Sample | Fluorescent Recognition Element | Ref. |
|---|---|---|---|
| Brucellosis | Bovine sera | Flu-OPS | [20,34,47,49] * |
| Milk | Flu-OPS | [35] | |
| Sheep sera | Flu-OPS | [17,36] | |
| Pig sera | Flu-OPS | [37] | |
| Cervid sera | Flu-OPS | [38] | |
| Camel sera | Flu-OPS | [46] | |
| Salmonellosis | Egg yolk Chicken sera | Flu-OPS | [21] [53] |
| Tuberculosis | Bovine sera | Flu-MPB70 | [21,39] |
| Bovine sera | Flu-ESAT-6 protein | [40] | |
| Bovine sera | Flu-peptide | [54,56] * | |
| Paratuberculosis | Bovine sera | Flu-protein fraction of 35 kDa from Map 3065 | [24] |
| Equine infectious anemia virus | Horse sera | Flu-peptide | [23,41,61] * |
| Influenza A virus | Chicken and goat sera | Flu-peptide | [58] |
| Chicken and goat sera | Recombinant H5 subtype HA Flu-proteins | [65] | |
| SARS-CoV-2 Antibody | Human sera | SARS-CoV-2 receptor binding domain (Flu-RBD) protein | [28] |
5. Detection of Virus Particles with the Use of FPA
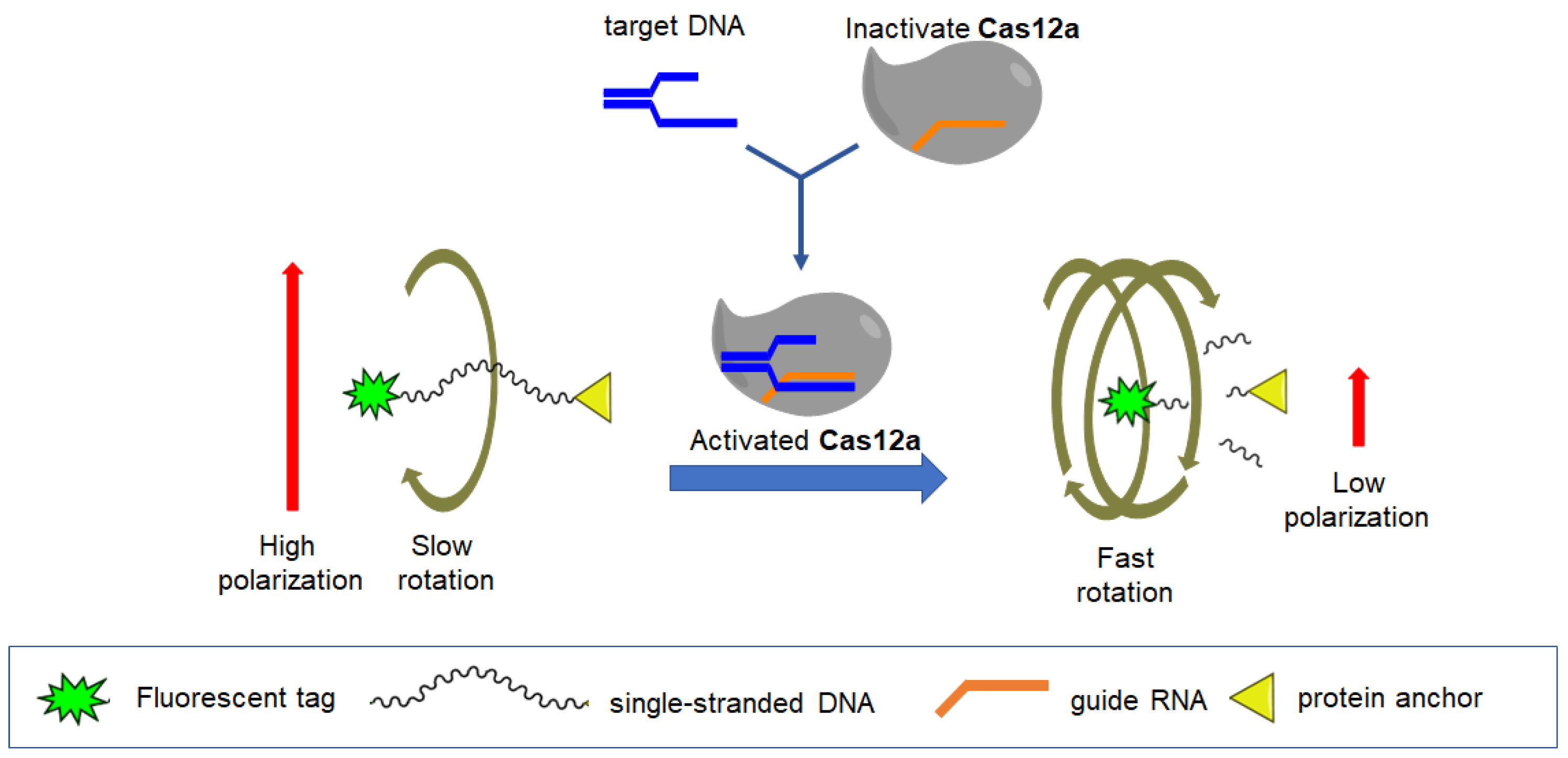
6. Detection of Nucleic Acid with the Use of FPA
7. Conclusions and Future Trends for the Use of FPA for Infection Detection
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- da Silva, R.E.; Novaes, M.R.C.G.; de Oliveira, C.; Guilhem, D.B. The impact of social cohesion and risk communication on excess mortality due to COVID-19 in 213 countries: A retrospective analysis. BMC Public Health 2024, 24, 1598. [Google Scholar] [CrossRef]
- Agunos, A.; Pierson, F.W.; Lungu, B.; Dunn, P.A.; Tablante, N. Review of Nonfoodborne Zoonotic and Potentially Zoonotic Poultry Diseases. Avian Dis. 2016, 60, 553–575. [Google Scholar] [CrossRef]
- Zumla, A.; Rao, M.; Wallis, R.S.; Kaufmann, S.H.E.; Rustomjee, R.; Mwaba, P.; Vilaplana, C.; Yeboah-Manu, D.; Chakaya, J.; Ippolito, G.; et al. Host-Directed Therapies Network consortium. Host-directed therapies for infectious diseases: Current status, recent progress, and future prospects. Lancet Infect. Dis. 2016, 16, e47–e63. [Google Scholar] [CrossRef]
- Walker, D.H. Principles of Diagnosis of Infectious Diseases. Pathobiol. Hum. Dis. 2014, 222–225. [Google Scholar] [CrossRef]
- Tram, D.T.N.; Wang, H.; Sugiarto, S.; Li, T.; Ang, W.H.; Lee, C.; Pastorin, G. Advances in nanomaterials and their applications in point of care (POC) devices for the diagnosis of infectious diseases. Biotechnol. Adv. 2016, 34, 1275–1288. [Google Scholar] [CrossRef]
- Alles, S.; Roman, B.; Le, Q.-N.; Kurteu, M.; Elmerhebi, E.; Potter, C.; Mozola, M.; Thompson, W.; Bastin, B.; Donofrio, R. Validation of the One Broth One Plate for Salmonella Method for Detection of Salmonella Spp. in Select Food and Environmental Samples: AOAC Performance Tested MethodSM 102002. J. AOAC Int. 2021, 104, 765–775. [Google Scholar] [CrossRef]
- Ramanan, P.; Bryson, A.L.; Binnicker, M.J.; Pritt, B.S.; Patel, R. Syndromic panel-based testing in clinical microbiology. Clin. Microbiol. Rev. 2018, 31, e00024-17. [Google Scholar] [CrossRef]
- Tang, Y.J.; Li, B.; Dai, J.G.; Dai, J.F.; Wang, X.H.; Si, J.; Ali, Z.; Li, T.T.; He, N.Y. Genotyping of Pseudomonas aeruginosa type III secretion system using magnetic enrichment multiplex polymerase chain reaction and chemiluminescence. J. Biomed. Nanotechnol. 2016, 12, 762–769. [Google Scholar] [CrossRef]
- Park, K.S.; Charles, R.C.; Ryan, E.T.; Weissleder, R.; Lee, H. Fluorescence Polarization Based Nucleic Acid Testing for Rapid and Cost-Effective Diagnosis of Infectious Disease. Chem.-Eur. J. 2015, 21, 16359–16363. [Google Scholar] [CrossRef]
- Wellinghausen, N.; Abele-Horn, M.; Donoso Mantke, O.; Enders, M.; Fingerle, V.; Gärtner, B.; Hagedorn, J.; Rabenau, H.F.; Reiter-Owona, I.; Tintelnot, K.; et al. MiQ Immunological Methods for the Detection of Infectious Diseases; Dustri-Verlag Dr. Karl Feistle: Munich, Germany, 2016. [Google Scholar]
- Praud, A.; Durán-Ferrer, M.; Fretin, D.; Jaÿ, M.; O’Connor, M.; Stournara, A.; Tittarelli, M.; Travassos, D.I.; Garin-Bastuji, B. Evaluation of three competitive ELISAs and a fluorescence polarization assay for the diagnosis of bovine brucellosis. Vet J. 2016, 216, 38–44. [Google Scholar] [CrossRef]
- Lee, H.; Kim, S.I. Review of Liquid Chromatography-Mass Spectrometry-Based Proteomic Analyses of Body Fluids to Diagnose Infectious Diseases. Int. J. Mol. Sci. 2022, 23, 2187. [Google Scholar] [CrossRef]
- Hull-Jackson, C.; Glass, M.B.; Ari, M.D.; Bragg, S.L.; Branch, S.L.; Whittington, C.U.; Edwards, C.N.; Levett, P.N. Evaluation of a commercial latex agglutination assay for serological diagnosis of leptospirosis. J. Clin. Microbiol. 2006, 44, 1853–1855. [Google Scholar] [CrossRef]
- Memish, Z.A.; Almuneef, M.; Mah, M.W.; Qassem, L.A.; Osoba, A.O. Comparison of the Brucella Standard Agglutination Test with the ELISA IgG and IgM in patients with Brucella bacteremia. Diagn. Microbiol. Infect. Dis. 2002, 44, 129–132. [Google Scholar] [CrossRef]
- Jolley, M.E.; Nasir, M.S. The use of fluorescence polarization assays for the detection of infectious diseases. Comb. Chem. High Throughput Screen. 2003, 6, 235–244. [Google Scholar] [CrossRef]
- Sánchez-Villalobos, A.; Urdaneta-Fernández, M.; Rubio-Fuenmayor, E.; Molero-Saras, G.; Luzardo-Charris, C.; Corona-Mengual, C. Desarrollo y evaluación de un protocolo serológico de fluorescencia polarizada en el estudio de anticuerpos contra Brucella spp en humanos [Development and evaluation of a serological protocol of fluorescence polarization for the preliminary study of Brucella spp antibodies in humans]. Investig. Clin. 2011, 52, 48–57. [Google Scholar] [PubMed]
- Stournara, A.; Minas, A.; Bourtzi-Chatzopoulou, E.; Stack, J.; Koptopoulos, G.; Petridou, E.; Sarris, K. Assessment of serological response of young and adult sheep to conjunctival vaccination with Rev-1 vaccine by fluorescence polarization assay (FPA) and other serological tests for B. melitensis. Vet. Microbiol. 2007, 119, 53–64. [Google Scholar] [CrossRef]
- Mukhametova, L.I.; Eremin, S.A. Fluorescence polarization immunoassays of organic compounds for food safety. Modern Biotechnology and Food Safety Detection. Front. Biosci. 2024, 16, 4. [Google Scholar] [CrossRef]
- Camacho, R.; Täuber, D.; Scheblykin, I.G. Fluorescence Anisotropy Reloaded-Emerging Polarization Microscopy Methods for Assessing Chromophores’ Organization and Excitation Energy Transfer in Single Molecules, Particles, Films, and Beyond. Adv. Mater. 2019, 31, 1805671. [Google Scholar] [CrossRef]
- Marcelo, I.; Hernán, B.; Ruth, S.; Milena, G.; Judith, G.; Julio, P.; David, H.; Jorge, M.; Martín, C.; Byron, P. Determining a Diagnostic Cut-Off on Fluorescence Polarization Assay (FPA) for Bovine Brucellosis in Carchi, Ecuador. Open J. Anim. Sci. 2017, 07, 425–432. [Google Scholar] [CrossRef]
- Surujballi, O.M.; Lutze-Wallace, C.; Turcotte, C.; Savic, M.; Stevenson, D.; Romanowska, A.; Monagle, W.; Berlie-Surujballi, G.; Tangorra, E. Sensitive diagnosis of bovine tuberculosis in a farmed cervid herd with use of an MPB70 protein fluorescence polarization assay. Canad. J. Vet. Res. 2009, 73, 171–176. [Google Scholar] [PubMed]
- Gast, R.K.; Nasir, M.S.; Jolley, M.E.; Holt, P.S.; Stone, H.D. Detection of experimental Salmonella enteritidis and S. typhimurium infections in laying hens by fluorescence polarization assay for egg yolk antibodies. Poult. Sci. 2002, 81, 1128–1131. [Google Scholar] [CrossRef]
- Tencza, S.B.; Islam, K.R.; Kalia, V.; Nasir, M.S.; Jolley, M.E.; Montelaro, R.C. Development of a fluorescence polarization-based diagnostic assay for equine infectious anemia virus. J. Clin. Microbiol. 2000, 38, 1854–1859. [Google Scholar] [CrossRef]
- Torres-Velez, R.; Santillán-Flores, M.A.; Cordova-López, D.; Martinez-Martinez, O.L.; Guzmán-Ruiz, C.C. Comparison of fluorescence polarization assay and enzyme-linked immunosorbent assay for the diagnosis of bovine paratuberculosis. J. Vet. Med. Anim. Health 2019, 11, 94–99. [Google Scholar] [CrossRef]
- Jameson, D.M. Introduction to Fluorescence; Taylor & Francis: Boca Raton, FL, USA, 2014. [Google Scholar]
- Nishiyama, K.; Fukuyama, M.; Maeki, M.; Ishida, A.; Tani, H.; Hibara, A.; Tokeshi, M. One-step non-competitive fluorescence polarization immunoassay based on a Fab fragment for C-reactive protein quantification. Sens. Actuators B Chem. 2021, 326, 128982. [Google Scholar] [CrossRef]
- Nishiyama, K.; Takeda, Y.; Takahash, K.; Fukuyama, M.; Maeki, M.; Ishida, A.; Tani, H.; Shigemura, K.; Hibara, A.; Ogawa, H.; et al. Non-competitive fluorescence polarization immunoassay for detection of H5 avian influenza virus using a portable analyzer. Anal. Bioanal. Chem. 2021, 413, 4619–4623. [Google Scholar] [CrossRef]
- Nishiyama, K.; Takahashi, K.; Fukuyama, M.; Kasuya, M.; Imai, A.; Usukura, T.; Maishi, N.; Maeki, M.; Ishida, A.; Tani, H.; et al. Facile and rapid detection of SARS-CoV-2 antibody based on a noncompetitive fluorescence polarization immunoassay in human serum samples. Biosens. Bioelectron. 2021, 190, 113414. [Google Scholar] [CrossRef]
- Reid, C.W.; Fulton, K.M.; Twine, S.M. Never take candy from a stranger: The role of the bacterial glycome in host-pathogen interactions. Future Microbiol. 2010, 5, 267–288. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Kalscheuer, R.; Palacios, A.; Anso, I.; Cifuente, J.; Anguita, J.; Jacobs, W.R., Jr.; Guerin, M.E.; Prados-Rosales, R. The Mycobacterium tuberculosis capsule: A cell structure with key implications in pathogenesis. Biochem. J. 2019, 476, 1995–2016. [Google Scholar] [CrossRef]
- Daffé, M.; Etienne, G. The capsule of Mycobacterium tuberculosis and its implications for pathogenicity. Tuber. Lung Dis. 1999, 79, 153–169. [Google Scholar] [CrossRef]
- Bose, S.; Song, M.Y.; Nam, J.K.; Lee, M.J.; Kim, H. In vitro and in vivo protective effects of fermented preparations of dietary herbs against lipopolysaccharide insult. Food Chem. 2012, 134, 758–765. [Google Scholar] [CrossRef]
- Matope, G.; Muma, J.B.; Toft, N.; Gori, E.; Lund, A.; Nielsen, K.; Skjerve, E. Evaluation of sensitivity and specificity of RBT, c-ELISA and fluorescence polarization assay for diagnosis of brucellosis in cattle using latent class analysis. Vet. Immunol. Immunopathol. 2011, 141, 58–63. [Google Scholar] [CrossRef]
- Sotolongo-Rodríguez, D.; Gomez-Flores, R.; Navarro-Soto, M.C.; Arellano-Reynoso, B.; Tamez-Guerra, P.; Ramírez-Pfeiffer, C. Evaluation of the Fluorescence Polarization Assay for the Diagnosis of Brucellosis in Goat Milk. Vet. Sci. 2022, 9, 303. [Google Scholar] [CrossRef]
- Minas, A.; Stournara, A.; Minas, M.; Papaioannou, A.; Krikelis, V.; Tselepidis, S. Validation of fluorescence polarization assay (FPA) and comparison with other tests used for diagnosis of B. melitensis infection in sheep. Vet. Microbiol. 2005, 111, 211–221. [Google Scholar] [CrossRef]
- Olsen, S.C.; Tatum, F.M. Swine brucellosis: Current perspectives. Vet. Med. 2016, 8, 1–12. [Google Scholar] [CrossRef]
- Gall, D.; Nielsen, K.; Forbes, L.; Cook, W.; Leclair, D.; Balsevicius, S.; Kelly, L.; Smith, P.; Mallory, M. Evaluation of the fluorescence polarization assay and comparison to other serological assays for detection of brucellosis in cervids. J. Wildl. Dis. 2001, 37, 110–118. [Google Scholar] [CrossRef]
- Surujballi, O.P.; Romanowska, A.; Sugden, E.A.; Turcotte, C.; Jolley, M.E. A fluorescence polarization assay for the detection of antibodies to Mycobacterium bovis in cattle sera. Vet. Microbiol. 2002, 87, 149–157. [Google Scholar] [CrossRef]
- Javed, R.; Narang, D.; Kaur, P.; Chandra, M.; Filia, G.; Singh, S.T. A fluorescence polarization assay using recombinant protein ESAT-6 for the detection of antibodies against pathogenic Mycobacterium bovis in bovine. Iran. J. Vet. Res. 2022, 23, 204–209. [Google Scholar] [CrossRef]
- Patra, S.; Tellapragada, C.; Vandana, K.E.; Mukhopadhyay, C. Diagnostic utility of in-house loop-mediated isothermal amplification and real-time PCR targeting virB gene for direct detection of Brucella melitensis from clinical specimens. J. Appl. Microbiol. 2019, 27, 230–236. [Google Scholar] [CrossRef]
- Xiao, Y.; Zou, G.; Yin, J.; Tan, W.; Zhou, J.; Zhang, H. Seroepidemiology of human Brucella infection in Yixing, China. Trop. Dr. 2017, 47, 165–167. [Google Scholar] [CrossRef]
- Corbel, M.J. The relationship between the protective and cross-reacting antigens of Brucella spp., Yersinia enterocolitica 0:9 and Salmonella serotypes of Kaufmann-White group N. Contrib. Microbiol. Immunol. 1979, 5, 50–63. [Google Scholar] [PubMed]
- Nielsen, K.; Yu, W.L. Serological diagnosis of brucellosis. Prilozi 2010, 31, 65–89. [Google Scholar] [PubMed]
- Wang, Y.; Vallée, E.; Compton, C.; Heuer, C.; Guo, A.; Wang, Y.; Zhang, Z.; Vignes, M. A novel Bayesian Latent Class Model (BLCM) evaluates multiple continuous and binary tests: A case study for Brucella abortus in dairy cattle. Prev. Vet. Med. 2024, 224, 106115. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.-B.; Xiao, D.; Liu, J.-Y.; Bi, H.-M.; Zheng, Z.-R.; Wang, L.-D.; Yang, X.-W.; Tian, G.-Z.; Zhao, H.-Y.; Piao, D.-R.; et al. Fluorescence polarization assay improves the rapid detection of human brucellosis in China. Infect. Dis. Poverty 2021, 10, 46. [Google Scholar] [CrossRef]
- Gwida, M.M.; ElGohary, A.H.; Melzer, F.; Tomaso, H.; Rosler, U.; Wernery, U.; Wernery, R.; Elschner, M.C.; Khan, I.; Eickhoff, M.; et al. Comparison of diagnostic tests for the detection of Brucella spp. in camel sera. BMC Res Notes 2011, 4, 525. [Google Scholar] [CrossRef]
- Park, K.S.; Huang, C.H.; Lee, K.; Yoo, Y.E.; Castro, C.M.; Weissleder, R.; Lee, H. Rapid Identification of Health Care-Associated Infections with an Integrated Fluorescence Anisotropy System. Sci. Adv. 2016, 2, e1600300. [Google Scholar] [CrossRef] [PubMed]
- The Brucella FPA Test. Available online: https://ellielab.com/brucella-fpa/ (accessed on 1 April 2024).
- Ibarra, M.; Campos, M.; Hernán, B.; Loor-Giler, A.; Chamorro, A.; Nuñez, L. Comparison of diagnostic tests for detecting bovine brucellosis in animals vaccinated with S19 and RB51 strain vaccines. Vet. World 2023, 16, 2080–2085. [Google Scholar] [CrossRef]
- Andrade, R.S.; de Oliveira, M.M.; de Sousa Bueno Filho, J.S.; Ferreira, F.; Godfroid, J.; Lage, A.P.; Dorneles, E.M.S. Accuracy of serological tests for bovine brucellosis: A systematic review and meta-analysis. Prev. Vet. Med. 2023, 222, 106079. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, Y.; Yao, L.; Shang, H.; Zheng, Z.; Chen, W.; Xu, J. Self-Assembly of Multivalent Aptamer-Tethered DNA Monolayers Dedicated to a Fluorescence Polarization-Responsive Circular Isothermal Strand Displacement Amplification for Salmonella Assay. Anal. Chem. 2023, 95, 2570–2578. [Google Scholar] [CrossRef]
- Gast, R.K.; Nasir, M.S.; Jolley, M.E.; Holt, P.S.; Stone, H.D. Serological detection of experimental Salmonella enteritidis infections in laying hens by fluorescence polarization and enzyme immunoassay. Avian Dis. 2002, 46, 137–142. [Google Scholar] [CrossRef]
- Jolley, M.E.; Nasir, M.S.; Surujballi, O.P.; Romanowska, A.; Renteria, T.B.; De la Mora, A.; Lim, A.; Bolin, S.R.; Michel, A.L.; Kostovic, M.; et al. Fluorescence polarization assay for the detection of antibodies to Mycobacterium bovis in bovine sera. Vet. Microbiol. 2007, 120, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Wiker, H.G. MPB70 and MPB83--major antigens of Mycobacterium bovis. Scand. J. Immunol. 2009, 69, 492–499. [Google Scholar] [CrossRef] [PubMed]
- A Bovine Tuberculosis Antibody Test Kit Based on Fluorescence Polarization. Available online: https://ellielab.com/tb-fpa/ (accessed on 1 April 2024).
- Espasandin, A.G.; Cipolini, M.F.; Forletti, A.; Díaz, S.; Soto, J.; Martínez, D.E.; Storani, C.A.; Monzón, N.M.; Beltrame, J.I.; Lucchesi, E.; et al. Comparison of serological techniques for the diagnosis of equine infectious Anemia in an endemic area of Argentina. J. Virol. Methods 2021, 291, 114101. [Google Scholar] [CrossRef] [PubMed]
- da Cunha Beltrão, M.; de Siqueira Alencar, C.; Leite, A.; Freitas, L.; Gonzalez, J.; de Assis Santana, V.; Manso-Filho, H. Comparison of Two Protocols of Agar Gel Immunodiffusion (AGID) Used to Diagnose of Equine Infectious Anemia (EIA). Open J. Vet. Med. 2015, 5, 169–174. [Google Scholar] [CrossRef]
- Hu, Z.; Guo, K.; Du, C.; Sun, J.; Naletoski, I.; Chu, X.; Lin, Y.; Wang, X.; Barrandeguy, M.; Samuel, M.; et al. Development and evaluation of a blocking ELISA for serological diagnosis of equine infectious anemia. Appl. Microbiol. Biotechnol. 2023, 107, 3305–3317. [Google Scholar] [CrossRef] [PubMed]
- Nardini, R.; Autorino, G.L.; Ricci, I.; Frontoso, R.; Rosone, F.; Simula, M.; Scicluna, M.T. Validation according to OIE criteria of a monoclonal, recombinant p26-based, serologic competitive enzyme-linked immunosorbent assay as screening method in surveillance programs for the detection of Equine infectious anemia virus antibodies. J. Vet. Diagn. Investig. 2016, 28, 88–97. [Google Scholar] [CrossRef]
- Test Equine Infectious Anemia Accurately in the Field or Lab. Available online: https://ellielab.com/eia-fpa/ (accessed on 1 April 2024).
- Gortázar, C.; Barroso, P.; Nova, R.; Cáceres, G. The role of wildlife in the epidemiology and control of Foot-and-mouth-disease And Similar Transboundary (FAST) animal diseases: A review. Transbound. Emerg. Dis. 2022, 69, 2462–2473. [Google Scholar] [CrossRef]
- FAO Emergency Centre for Transboundary Animal Diseases (ECTAD). Available online: https://www.fao.org/animal-health/our-programmes/fao-emergency-centre-for-transboundary-animal-diseases-(ectad)/en (accessed on 1 April 2024).
- Takeda, Y.; Yonezawa, Y.; Asake, S.; Ogawa, H.; Imai, K. A fluorescence polarization immunoassay for the rapid detection of antibody against influenza A virus in chicken and goat sera. J. Vet. Diagn. Investig. 2020, 32, 887–891. [Google Scholar] [CrossRef]
- Nishiyama, K.; Takeda, Y.; Maeki, M.; Ishida, A.; Tani, H.; Shigemura, K.; Hibara, A.; Yonezawa, Y.; Imai, K.; Ogawa, H.; et al. Rapid detection of anti-H5 avian influenza virus antibody by fluorescence polarization immunoassay using a portable fluorescence polarization analyzer. Sens. Actuators B Chem. 2020, 316, 128160. [Google Scholar] [CrossRef]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef]
- Alandijany, T.A.; El-Kafrawy, S.A.; Tolah, A.M.; Sohrab, S.S.; Faizo, A.A.; Hassan, A.M.; Alsubhi, T.L.; Othman, N.A.; Azhar, E.I. Development and optimization of in-house ELISA for detection of human IgG antibody to SARS-CoV-2 full length spike protein. Pathogens 2020, 9, 803. [Google Scholar] [CrossRef] [PubMed]
- Roy, V.; Fischinger, S.; Atyeo, C.; Slein, M.; Loos, C.; Balazs, A.; Luedemann, C.; Astudillo, M.G.; Yang, D.; Wesemann, D.R.; et al. SARS-CoV-2-specific ELISA development. J. Immunol. Methods 2020, 484–485, 112832. [Google Scholar] [CrossRef]
- Okamatsu, M.; Feng, F.; Ohyanagi, T.; Nagahori, N.; Someya, K.; Sakoda, Y.; Miura, N.; Nishimura, S.; Kida, H. Fluorescence polarization-based assay using N-glycan-conjugated quantum dots for screening in hemagglutinin blockers for influenza A viruses. J. Virol. Methods 2013, 187, 390–394. [Google Scholar] [CrossRef]
- Zhao, Q.; Tao, J.; Uppal, J.S.; Peng, H.; Wang, H.; Le, X.C. Nucleic acid aptamers improving fluorescence anisotropy and fluorescence polarization assays for small molecules. TrAC Trends Anal. Chem. 2019, 110, 401–409. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, P.; Liu, T.; Pu, H.; Sun, D.-W. A fluorescence biosensor based on single-stranded DNA and carbon quantum dots for acrylamide detection. Food Chem. 2021, 356, 129668–129676. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ong, S.; Chen, Y.; Jimmy Huang, P.-J.; Liu, J. Label-free and Dye-free Fluorescent Sensing of Tetracyclines Using a Capture-Selected DNA Aptamer. Anal. Chem. 2022, 94, 10175–10182. [Google Scholar] [CrossRef] [PubMed]
- Gleerup, J.L.; Mogensen, T.H. CRISPR-Cas in Diagnostics and Therapy of Infectious Diseases. J. Infect. Dis. 2022, 226, 1867–1876. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyu, Y.; Wang, D.; Feng, M.; Shen, S.; Zhu, L.; Pan, C.; Zai, X.; Wang, S.; Guo, Y.; et al. Rapid Identification of Brucella Genus and Species In Silico and On-Site Using Novel Probes with CRISPR/Cas12a. Microorganisms 2024, 12, 1018. [Google Scholar] [CrossRef]
- Lee, C.Y.; Degani, I.; Cheong, J.; Lee, J.H.; Choi, H.J.; Cheon, J.; Lee, H. Fluorescence polarization system for rapid COVID-19 diagnosis. Biosens. Bioelectron. 2021, 178, 113049. [Google Scholar] [CrossRef]
- Marx, V. PCR heads into the field. Nat. Methods 2015, 12, 393–397. [Google Scholar] [CrossRef]
- Bonetta, L. Prime time for real-time PCR. Nat. Methods 2005, 2, 305–312. [Google Scholar] [CrossRef]
- Wang, L.; Tian, J.; Yang, W.; Zhao, Y.; Zhao, S. A T7exonuclease-assisted target recycling amplification with graphene oxide acting as the signal amplifier for fluorescence polarization detection of human immunodeficiency virus (HIV) DNA. Luminescence 2016, 31, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; He, G.; Tian, J.; Zhao, Y.; Zhao, S. Fluorescence polarization gene assay for HIV-DNA based on the use of dendrite-modified gold nanoparticles acting as signal amplifiers. Microchim. Acta 2018, 185, 119. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Rui, C.; Zhansheng, J.; Xiangling, W.; Ping, L.; Zhen, Y.; Yanhai, G.; Ju, Z. Detection of hepatitis B virus genotypes A to D by the fluorescence polarization assay based on asymmetric PCR. J. Virol. Methods 2010, 168, 31–37. [Google Scholar] [CrossRef]
- Feng, W.; Zhang, H.; Le, X.C. Signal Amplification by the trans-Cleavage Activity of CRISPR-Cas Systems: Kinetics and Performance. Anal. Chem. 2023, 95, 206–217. [Google Scholar] [CrossRef]
- Safenkova, I.V.; Samokhvalov, A.V.; Serebrennikova, K.V.; Eremin, S.A.; Zherdev, A.V.; Dzantiev, B.B. DNA Probes for Cas12a-Based Assay with Fluorescence Anisotropy Enhanced Due to Anchors and Salts. Biosensors 2023, 13, 1034. [Google Scholar] [CrossRef]
- Wang, L.; Wang, R.; Wei, H.; Li, Y. Selection of aptamers against pathogenic bacteria and their diagnostics application. World J. Microbiol. Biotechnol. 2018, 34, 149. [Google Scholar] [CrossRef]
- Ma, P.; Duan, N.; Ye, H.; Xia, Y.; Ding, Z.; Wang, Z. Selection, truncation and fluorescence polarization based aptasensor for Weissella viridescens detection. Talanta 2022, 246, 123499. [Google Scholar] [CrossRef]
- Argunov, D.A.; Krylov, V.B.; Nifantiev, N.E. Convergent synthesis of isomeric heterosaccharides related to the fragments of galactomannan from Aspergillus fumigatus. Org. Biomol. Chem. 2015, 13, 3255–3267. [Google Scholar] [CrossRef]
- Gening, M.L.; Tsvetkov, Y.E.; Pier, G.B.; Nifantiev, N.E. Synthesis of oligo-β(1→6)-glucosamines corresponding to the fragments of the surface polysaccharide of Staphylococcus aureus. Carbohydr. Res. 2007, 342, 567–575. [Google Scholar] [CrossRef]
- Komarova, B.K.; Wong, S.S.; Orekhova, M.V.; Tsvetkov, Y.E.; Krylov, V.B.; Beauvais, A.; Bouchara, J.-P.; Kearney, J.; Aimanianda, V.; Latgé, J.-P.; et al. Chemical synthesis and application of biotinylated oligo-α-(1→3)-D-glucosides to study the antibody and cytokine response against the cell wall α-(1→3)-D-glucan of Aspergillus fumigatus. J. Org. Chem. 2018, 83, 12965–12976. [Google Scholar] [CrossRef] [PubMed]
- Kazakova, E.D.; Yashunsky, D.V.; Krylov, V.B.; Bouchara, J.-P.; Cornet, M.; Valsecchi, I.; Fontaine, T.; Latgé, J.-P.; Nifantiev, N.E. Biotinylated oligo-α-(1→4)-D-galactosamines and their N-acetylated derivatives: α-stereoselective synthesis and immunology application. J. Am. Chem. Soc. 2020, 142, 1175–1179. [Google Scholar] [CrossRef] [PubMed]
- Laverde, D.; Romero-Saavedra, F.; Argunov, D.A.; Enotarpi, J.; Krylov, V.B.; Kalfopoulou, E.; Martini, R.; Torelli, C.; van der Marel, G.A.; Sanguinetti, M.; et al. Synthetic Oligomers Mimicking Capsular Polysaccharide Diheteroglycan are Potential Vaccine Candidates against Encapsulated Enterococcal Infections. ACS Infect. Dis. 2020, 6, 1816–1826. [Google Scholar] [CrossRef]
- Dorokhova, V.S.; Gerbst, A.G.; Komarova, B.S.; Previato, J.O.; Mendonça Previato, L.; Dmitrenok, A.S.; Shashkov, A.S.; Krylov, V.B.; Nifantiev, N.E. Synthesis and conformational analysis of vicinally branched trisaccharide β-D-Galf-(1→2)-[β-D-Galf-(1→3)-]-α-Galp from Cryptococcus neoformans galactoxylomannan. Org. Biomol. Chem. 2021, 19, 2923–2931. [Google Scholar] [CrossRef] [PubMed]
- Solovev, A.S.; Denisova, E.M.; Kurbatova, E.A.; Kutsevalova, O.Y.; Boronina, L.G.; Ageevets, V.A.; Sidorenko, S.V.; Krylov, V.B.; Nifantiev, N.E. Synthesis of methylphosphorylated oligomannosides structurally related to lipopolysaccharide O-antigens of Klebsiella pneumoniae serotype O3 and their application for detection of specific antibodies in rabbit and human sera. Org. Biomol. Chem. 2023, 21, 8306–8319. [Google Scholar] [CrossRef] [PubMed]
- Krylov, V.B.; Petruk, M.I.; Karimova, M.R.; Mukhametova, L.I.; Matveev, A.L.; Tikunova, N.V.; Eremin, S.A.; Nifantiev, N.E. Potential of polarization fluorescence immunoassay for the determination of galactomannan of Aspergillus fumigatus. Russ. Chem. Bull. Int. Ed. 2019, 68, 2365–2369. [Google Scholar] [CrossRef]
- Mukhametova, L.I.; Krylov, V.B.; Solovev, A.S.; Yashunsky, D.V.; Matveev, A.L.; Tikunova, N.V.; Eremin, S.A.; Nifantiev, N.E. Affinity characteristics of anti-β-(1→3)-D-glucan monoclonal antibody 3G11 by fluorescence polarization immunoassay. Russ. Chem. Bull. 2021, 70, 975–981. [Google Scholar] [CrossRef]
- Mukhametova, L.I.; Zherdev, D.O.; Kuznetsov, A.N.; Yudina, O.N.; Tsvetkov, Y.E.; Eremin, S.A.; Krylov, V.B.; Nifantiev, N.E. Fluorescence-Polarization-Based Assaying of Lysozyme with Chitooligosaccharide Tracers. Biomolecules 2024, 14, 170. [Google Scholar] [CrossRef]
- Mukhametova, L.I.; Zherdev, D.O.; Eremin, S.A.; Kuznetsov, A.N.; Yudin, V.I.; Sclyarov, O.D.; Babicheva, O.V.; Motorygin, A.V.; Tsvetkov, Y.E.; Krylov, V.B.; et al. Applying a fluorescence polarization assay for detection of brucellosis in animals using the fluorescently labeled synthetic oligosaccharides as biosensing tracer. Biosensors 2024, 14, 404. [Google Scholar] [CrossRef]
- Tsvetkov, Y.E.; Volkov, T.M.; Eremin, S.A.; Sklyarov, O.D.; Kulakov, Y.K.; Krylov, V.B.; Nifantiev, N.E. New Synthesis of oligosaccharides modelling the M epitope of the Brucella O-polysaccharide. Front. Chem. 2024, 12, 1424157. [Google Scholar] [CrossRef]

| Infectious Agent | Se | Sp | Cut-Off (mP) | Ref. |
|---|---|---|---|---|
| B. abortus and B. melitensis | 88.7 99.0 | 92.5 95 | 89.9 90 | [20] [34] |
| B. melitensis | 95 | 100 | 74.1 | [35] |
| B. melitensis | 97.6 | 98.9 | 87 | [36] |
| B. suis | 93.5 | 97.2 | 84 | [37] |
| B. abortus and B. suis | 99 | 84 | 80 | [38] |
| M. bovis | 92.9 | 98.3 | 173.9 | [39] |
| 96.9 | 64.6 | 127 | [40] | |
| Paratuberculosis | 88.5 | 91.4 | 126 | [24] |
| Equine infectious anemia virus | 98 | 98 | 98.6 | [21] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eremin, S.A.; Mukhametova, L.I.; Krylov, V.B.; Nifantiev, N.E. Fluorescence Polarization Assay for Infection Diagnostics: A Review. Molecules 2024, 29, 4712. https://doi.org/10.3390/molecules29194712
Eremin SA, Mukhametova LI, Krylov VB, Nifantiev NE. Fluorescence Polarization Assay for Infection Diagnostics: A Review. Molecules. 2024; 29(19):4712. https://doi.org/10.3390/molecules29194712
Chicago/Turabian StyleEremin, Sergei A., Liliya I. Mukhametova, Vadim B. Krylov, and Nikolay E. Nifantiev. 2024. "Fluorescence Polarization Assay for Infection Diagnostics: A Review" Molecules 29, no. 19: 4712. https://doi.org/10.3390/molecules29194712
APA StyleEremin, S. A., Mukhametova, L. I., Krylov, V. B., & Nifantiev, N. E. (2024). Fluorescence Polarization Assay for Infection Diagnostics: A Review. Molecules, 29(19), 4712. https://doi.org/10.3390/molecules29194712








