A Spectroscopic and In Silico Description of the Non-Covalent Interactions of Phthalic Acid Imide Derivatives with Deoxyribonucleic Acid—Insights into Their Binding Characteristics and Potential Applications
Abstract
:1. Introduction
2. Results and Discussion
2.1. Spectroscopic Studies
2.1.1. UV-Vis Spectroscopy
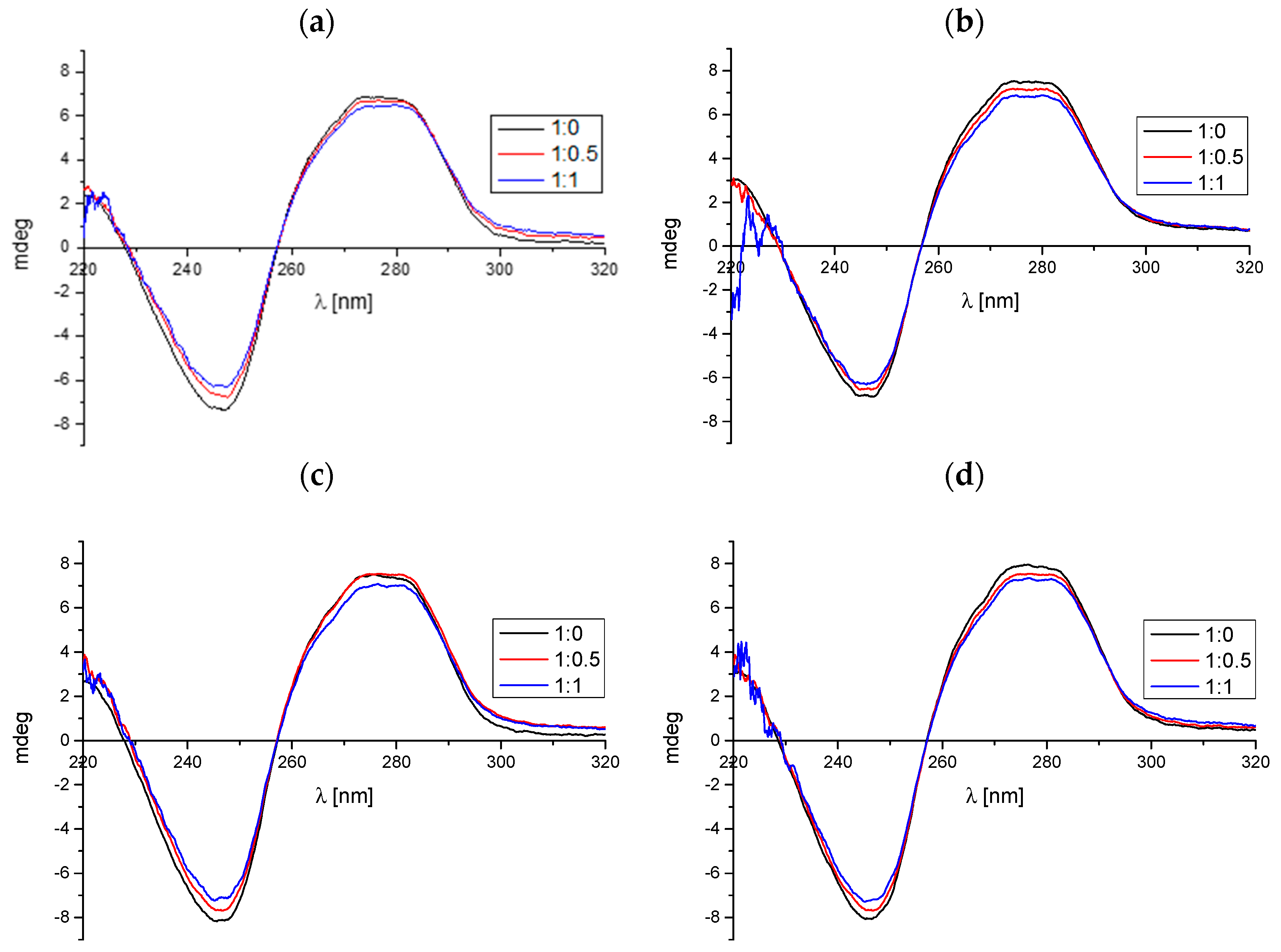
2.1.2. Circular Dichroism Spectroscopy
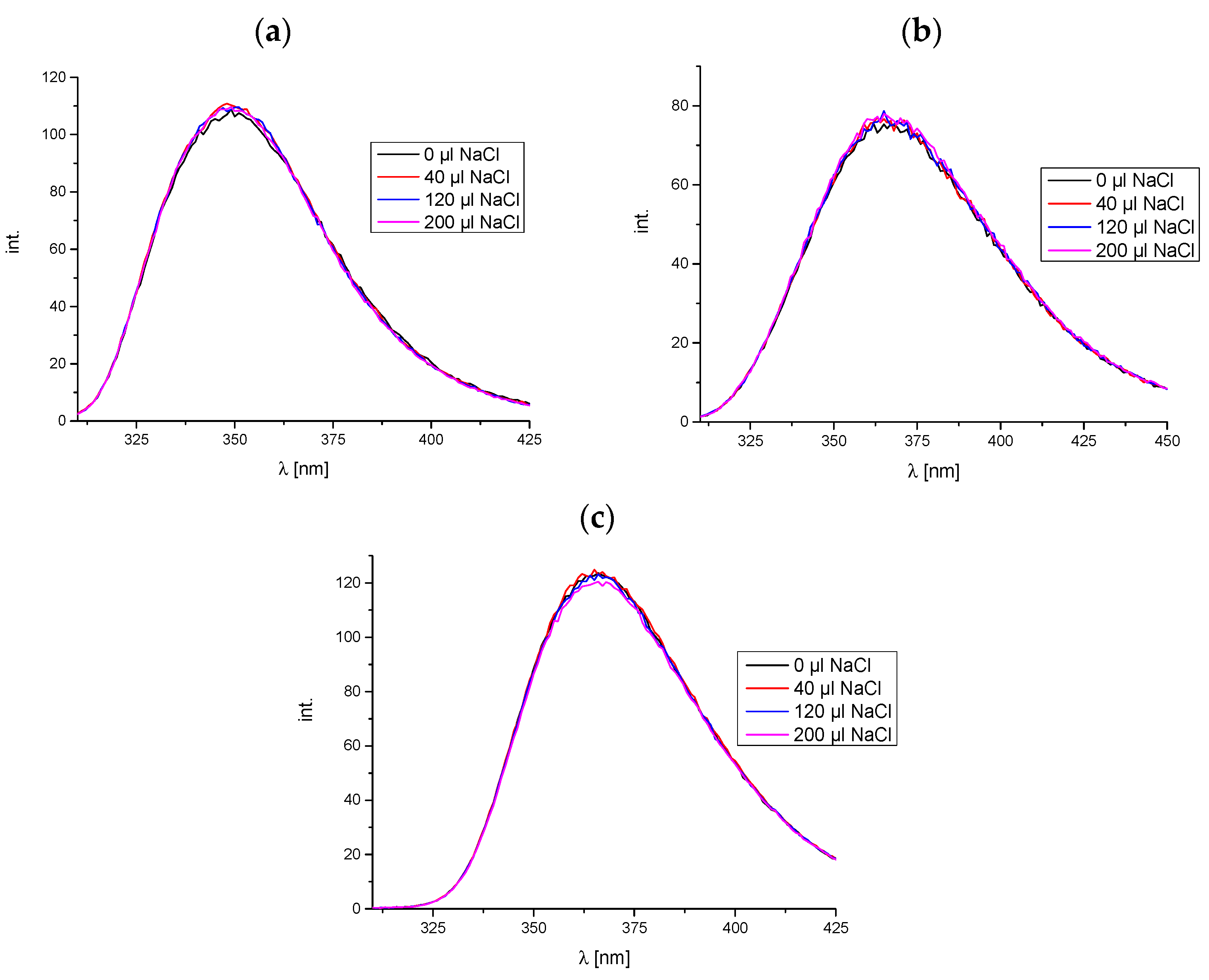
2.1.3. Fluorescence Spectroscopy
- Investigation of the occurrence of electrostatic interactions in the tested systems
- Competitive binding studies with markers
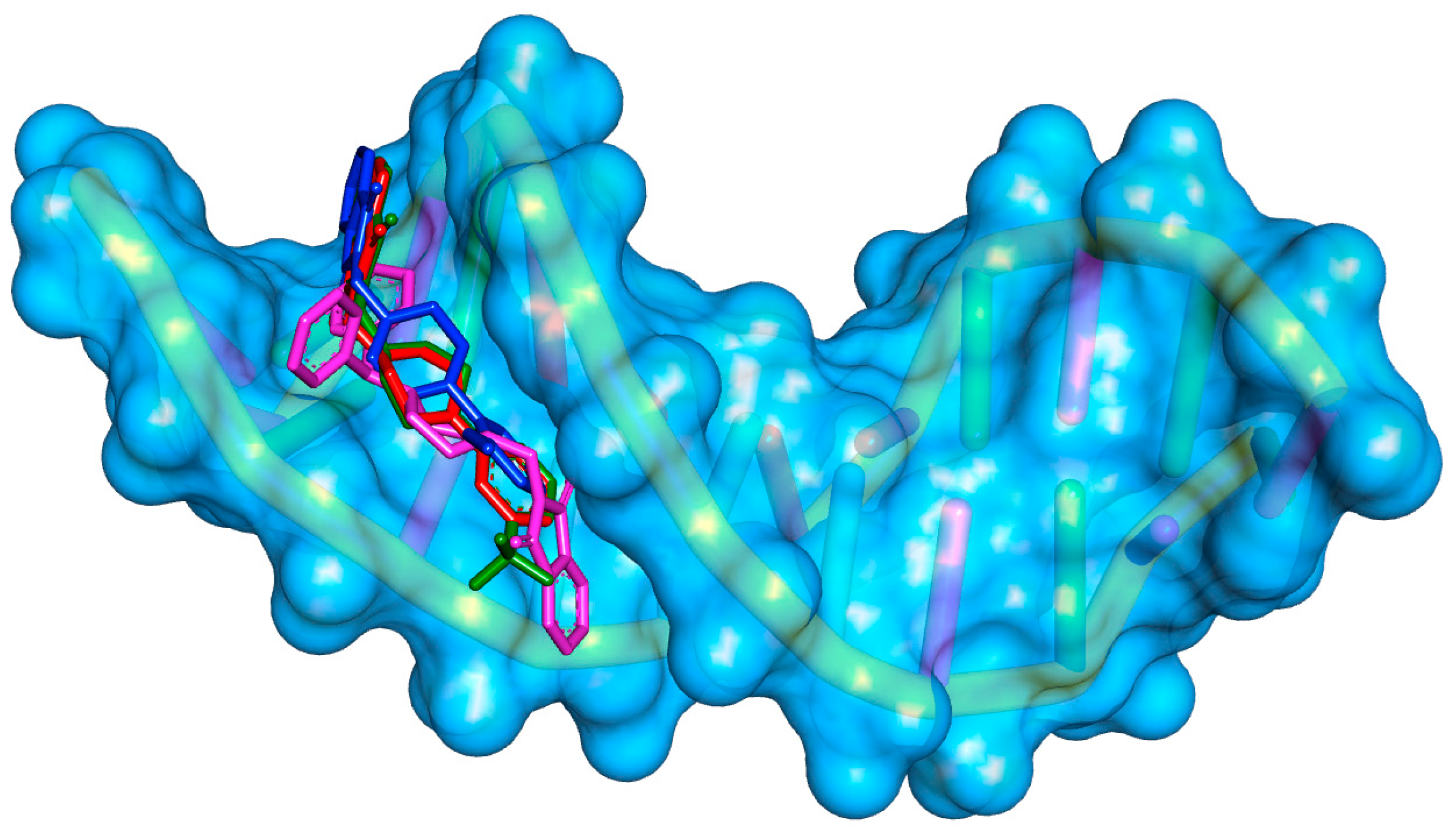
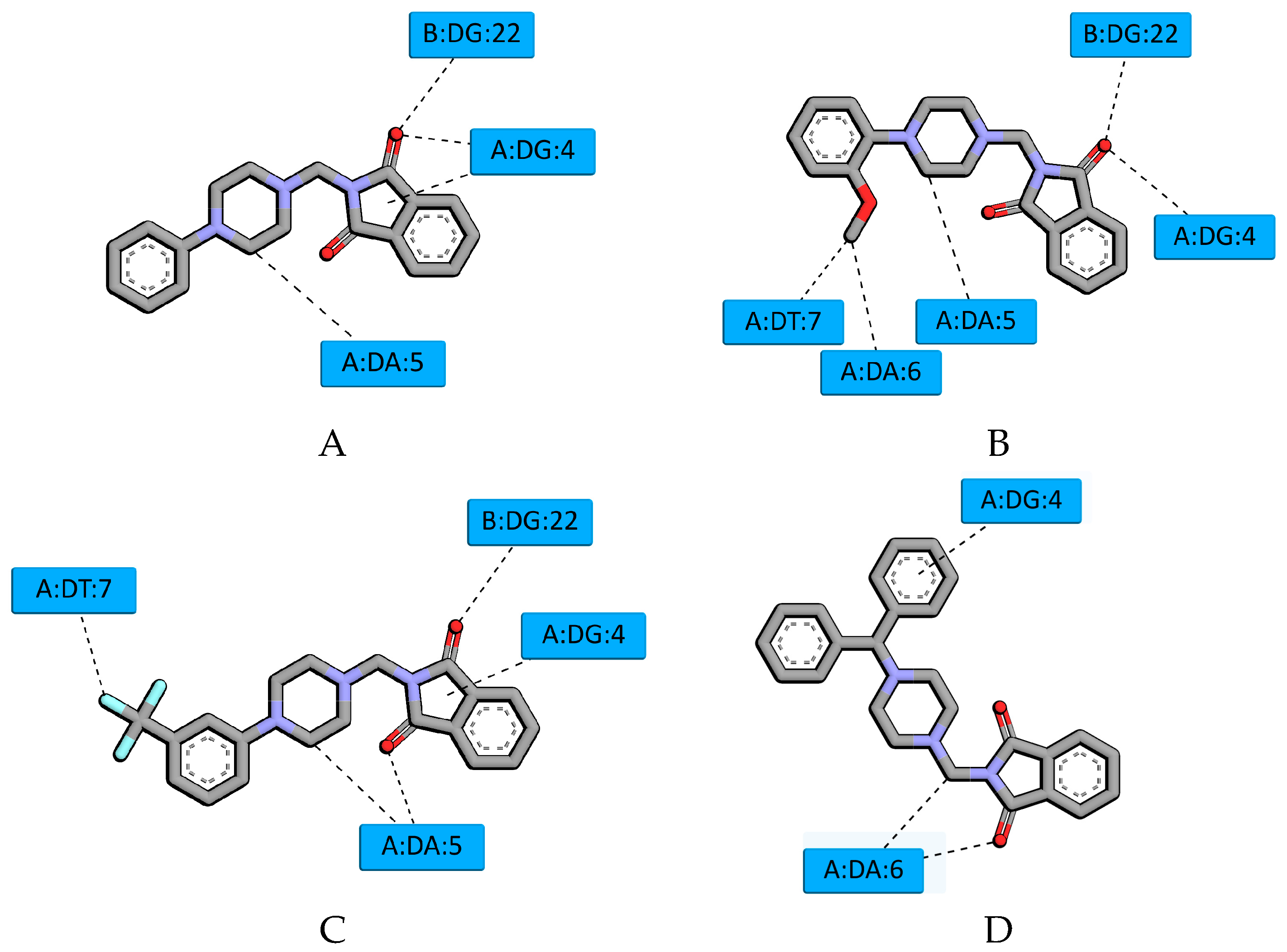
2.2. Molecular Modeling Study
3. Materials and Methods
3.1. Chemicals and Sample Preparation
3.2. Spectroscopic Methods
3.2.1. UV-Vis Spectroscopy
3.2.2. Circular Dichroism Spectroscopy
3.2.3. Fluorescence Spectroscopy
3.3. Molecular Modeling
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdulrahman, H.S.; Hassan Mohammed, M.; Al-Ani, L.A.; Ahmad, M.H.; Hashim, N.M.; Yehye, W.A. Synthesis of Phthalimide Imine Derivatives as a Potential Anticancer Agent. J. Chem. 2020, 2020, 3928204. [Google Scholar] [CrossRef]
- Bansal, M.; Upadhyay, C.; Poonam; Kumar, S.; Rathi, B. Phthalimide analogs for antimalarial drug discovery. RSC Med. Chem. 2021, 12, 1854–1867. [Google Scholar] [CrossRef] [PubMed]
- Krzyżak, E.; Kotynia, A.; Szkatuła, D.; Marciniak, A. Spectroscopic and Theoretical Analysis of the Interaction between Plasma Proteins and Phthalimide Analogs with Potential Medical Application. Life 2023, 13, 760. [Google Scholar] [CrossRef] [PubMed]
- Szkatuła, D.; Krzyżak, E.; Stanowska, P.; Duda, M.; Wiatrak, B. A new n-substituted 1h-isoindole-1,3(2h)-dione derivative—synthesis, structure and affinity for cyclooxygenase based on in vitro studies and molecular docking. Int. J. Mol. Sci. 2021, 22, 7678. [Google Scholar] [CrossRef]
- Dziubina, A.; Szkatuła, D.; Gdula-Argasińska, J.; Sapa, J. Synthesis and antinociceptive activity of four 1H-isoindolo-1,3(2H)-diones. Arch. Pharm. 2022, 355, 2100423. [Google Scholar] [CrossRef]
- Nayab, P.S.; Pulaganti, M.; Chitta, S.K.; Abid, M.; Uddin, R. Evaluation of DNA Binding, Radicals Scavenging and Antimicrobial Studies of Newly Synthesized N-Substituted Naphthalimides: Spectroscopic and Molecular Docking Investigations. J. Fluoresc. 2015, 25, 1905–1920. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Yasmeen, S.; Qais, F.A.; Rana, M.; Islam, A. Rahisuddin Binding and thermodynamic study of thalidomide with calf thymus DNA: Spectroscopic and computational approaches. Int. J. Biol. Macromol. 2022, 207, 644–655. [Google Scholar] [CrossRef]
- Pavithra, K.; Priyadharshini, R.D.; Vennila, K.N.; Elango, K.P. Exploring the binding mechanism of Pomalidomide drug with CT-DNA: Insights from multi-spectroscopic, molecular docking and simulation studies. J. Mol. Struct. 2024, 1302, 137547. [Google Scholar] [CrossRef]
- Xu, L.; Hu, Y.X.; Li, Y.C.; Zhang, L.; Ai, H.X.; Liu, Y.F.; Liu, H.S. In vitro DNA binding studies of lenalidomide using spectroscopic in combination with molecular docking techniques. J. Mol. Struct. 2018, 1154, 9–18. [Google Scholar] [CrossRef]
- Mukherjee, A.; Mondal, S.; Singh, B. Spectroscopic, electrochemical and molecular docking study of the binding interaction of a small molecule 5H-naptho [2,1-f][1,2] oxathieaphine 2,2-dioxide with calf thymus DNA. Int. J. Biol. Macromol. 2017, 101, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Shahabadi, N.; Maghsudi, M. Multi-spectroscopic and molecular modeling studies on the interaction of antihypertensive drug; Methyldopa with calf thymus DNA. Mol. Biosyst. 2014, 10, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Hu, Y.J.; Wang, H.; Yu, B.Q.; Yue, H.L. Molecular spectroscopy evidence of berberine binding to DNA: Comparative binding and thermodynamic profile of intercalation. Biomacromolecules 2012, 13, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Mandal, G.; Ganguly, T. Detailed spectroscopic investigations to reveal the nature of interaction of anionic porphyrin with calf thymus DNA. J. Photochem. Photobiol. B Biol. 2010, 101, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Devi, C.S.; Thulasiram, B.; Satyanarayana, S.; Nagababu, P. Analytical Techniques Used to Detect DNA Binding Modes of Ruthenium(II) Complexes with Extended Phenanthroline Ring. J. Fluoresc. 2017, 27, 2119–2130. [Google Scholar] [CrossRef]
- Benesi, H.A.; Hildebrand, J.H. A Spectrophotometric Investigation of the Interaction of Iodine with Aromatic Hydrocarbons. J. Am. Chem. Soc. 1949, 71, 2703–2707. [Google Scholar] [CrossRef]
- Chaires, J.B. Energetics of Drug—DNA. Biopolymers 1997, 44, 201–215. [Google Scholar] [CrossRef]
- Haq, I. Thermodynamics of drug-DNA interactions. Arch. Biochem. Biophys. 2002, 403, 1–15. [Google Scholar] [CrossRef]
- Jiménez, J.S.; Benítez, M.J. Gibbs Free Energy and Enthalpy–Entropy Compensation in Protein–Ligand Interactions. Biophysica 2024, 4, 298–309. [Google Scholar] [CrossRef]
- Uma, V.; Kanthimathi, M.; Weyhermuller, T.; Nair, B.U. Oxidative DNA cleavage mediated by a new copper (II) terpyridine complex: Crystal structure and DNA binding studies. J. Inorg. Biochem. 2005, 99, 2299–2307. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovská, I.; Renciuk, D.; Vorlícková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Nayab, P.S.; Pulaganti, M.; Chitta, S.K.; Oves, M. Rahisuddin Synthesis, spectroscopic studies of novel N-substituted phthalimides and evaluation of their antibacterial, antioxidant, DNA binding and molecular docking studies. Bangladesh J. Pharmacol. 2015, 10, 703–713. [Google Scholar] [CrossRef]
- Arjmand, F.; Jamsheera, A.; Mohapatra, D.K. Synthesis, characterization and in vitro DNA binding and cleavage studies of Cu(II)/Zn(II) dipeptide complexes. J. Photochem. Photobiol. B Biol. 2013, 121, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Karidi, K.; Garoufis, A.; Tsipis, A.; Hadjiliadis, N.; Den Dulk, H.; Reedijk, J. Synthesis, characterization, in vitro antitumor activity, DNA-binding properties and electronic structure (DFT) of the new complex cis-(Cl,Cl)[RuIICl2(NO+)(terpy)]Cl. Dalt. Trans. 2005, 7, 1176–1187. [Google Scholar] [CrossRef] [PubMed]
- Wani, T.A.; Alsaif, N.; Bakheit, A.H.; Zargar, S.; Al-Mehizia, A.A.; Khan, A.A. Interaction of an abiraterone with calf thymus DNA: Investigation with spectroscopic technique and modelling studies. Bioorg. Chem. 2020, 100, 103957. [Google Scholar] [CrossRef]
- Kapuscinski, J.; Darzynkiewicz, Z. Interactions of Acridine Orange with Double Stranded Nucleic Acids. Spectral and Affinity Studies. J. Biomol. Struct. Dyn. 1987, 5, 127–143. [Google Scholar] [CrossRef]
- Lyles, M.B.; Cameron, I.L. Interactions of the DNA intercalator acridine orange, with itself, with caffeine, and with double stranded DNA. Biophys. Chem. 2002, 96, 53–76. [Google Scholar] [CrossRef]
- de Oliveira, T.A.; Medaglia, L.R.; Maia, E.H.B.; Assis, L.C.; de Carvalho, P.B.; da Silva, A.M.; Taranto, A.G. Evaluation of Docking Machine Learning and Molecular Dynamics Methodologies for DNA-Ligand Systems. Pharmaceuticals 2022, 15, 132. [Google Scholar] [CrossRef]
- Huang, Z.Y.; Wang, Z.H.; Niu, Y.; Deng, G.X.; Bai, A.M.; Li, X.Y.; Hu, Y.J. Structure-dependent of 3-fluorooxindole derivatives interacting with ctDNA: Binding effects and molecular docking approaches. Bioorg. Chem. 2022, 121, 105698. [Google Scholar] [CrossRef]
- Mallena, S.; Lee, M.P.H.; Bailly, C.; Neidle, S.; Kumar, A.; Boykin, D.W.; Wilson, W.D. Thiophene-based diamidine forms a “super” AT binding minor groove agent. J. Am. Chem. Soc. 2004, 126, 13659–13669. [Google Scholar] [CrossRef]
- Mi, R.; Tu, B.; Bai, X.T.; Chen, J.; Ouyang, Y.; Hu, Y.J. Binding properties of palmatine to DNA: Spectroscopic and molecular modeling investigations. Luminescence 2015, 30, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Arafa, R.K.; Ismail, M.A.; Wenzler, T.; Brun, R.; Paul, A.; Wilson, W.D.; Alakhdar, A.A.; Boykin, D.W. New antiparasitic flexible triaryl diamidines, their prodrugs and aza analogues: Synthesis, in vitro and in vivo biological evaluation, and molecular modelling studies. Eur. J. Med. Chem. 2021, 222, 113625. [Google Scholar] [CrossRef] [PubMed]
- Szczęśniak-Sięga, B.M.; Zaręba, N.; Czyżnikowska, Ż.; Janek, T.; Kepinska, M. Rational Design, Synthesis, Molecular Docking, and Biological Evaluations of New Phenylpiperazine Derivatives of 1,2-Benzothiazine as Potential Anticancer Agents. Molecules 2024, 29, 4282. [Google Scholar] [CrossRef] [PubMed]
- Nafie, M.S.; Arafa, K.; Sedky, N.K.; Alakhdar, A.A.; Arafa, R.K. Triaryl dicationic DNA minor-groove binders with antioxidant activity display cytotoxicity and induce apoptosis in breast cancer. Chem. Biol. Interact. 2020, 324, 109087. [Google Scholar] [CrossRef] [PubMed]
- Kotynia, A.; Krzyżak, E.; Żądło, J.; Witczak, M.; Szczukowski, Ł.; Mucha, J.; Świątek, P.; Marciniak, A. Anti-Inflammatory and Antioxidant Pyrrolo[3,4-d]pyridazinone Derivatives Interact with DNA and Bind to Plasma Proteins—Spectroscopic and In Silico Studies. Int. J. Mol. Sci. 2024, 25, 1784. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208-IN1. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16. Revision C.01, Gaussian, Inc.: Wallingford, CT, USA, 2016. Available online: https://gaussian.com/citation/ (accessed on 29 October 2024).
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. Ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hitzenberger, M.; Rieger, M.; Kern, N.R.; Zacharias, M.; Im, W. CHARMM-GUI supports the Amber force fields. J. Chem. Phys. 2020, 153, 035103. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]

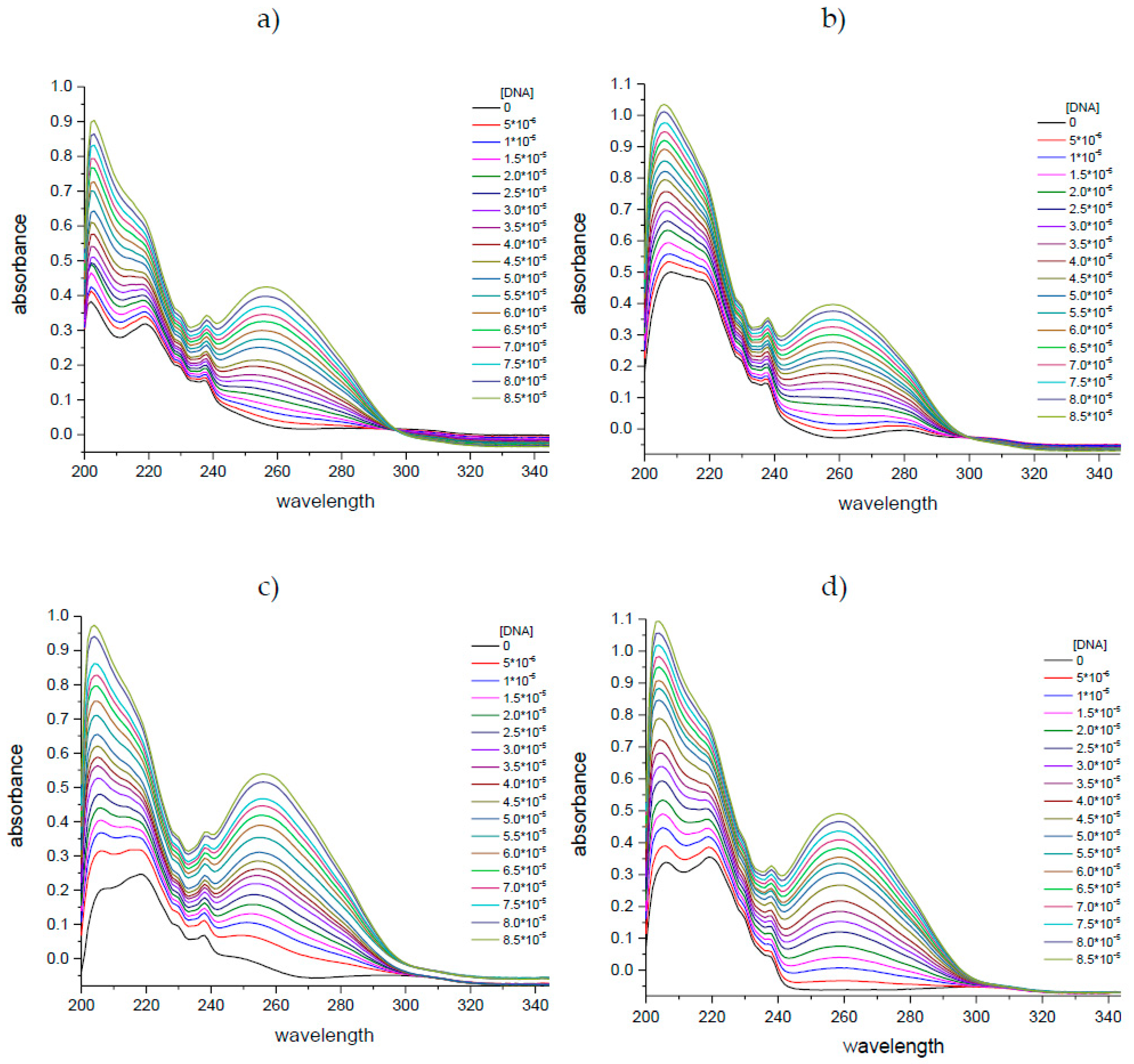



| Complex | Intercept | Slope | R2 |
|---|---|---|---|
| A/ctDNA | 0.3595 | 2×10−4 | 0.9802 |
| B/ctDNA | 0.2899 | 1×10−4 | 0.9941 |
| C/ctDNA | 0.8928 | 7×10−5 | 0.9899 |
| D/ctDNA | 0.3937 | 8×10−5 | 0.9987 |
| Complex | % Hyperchromicity | K [dm3·mol−1] | ΔG [J·mol−1] |
|---|---|---|---|
| A/ctDNA | 58.75 | 1.80×103 | −1.86×104 |
| B/ctDNA | 51.76 | 2.90×103 | −1.98×104 |
| C/ctDNA | 76.40 | 1.28×104 | −2.34×104 |
| D/ctDNA | 70.84 | 4.86×103 | −2.10×104 |
| Compound/ctDNA Molar Ratio | Δε245nm [mdeg] | Δε280nm [mdeg] |
|---|---|---|
| Compound A | ||
| 0 | −8.08 | 7.91 |
| 0.5 | −7.63 | 7.57 |
| 1 | −7.23 | 7.29 |
| Compound B | ||
| 0 | −6.83 | 7.52 |
| 0.5 | −6.50 | 7.12 |
| 1 | −6.25 | 6.87 |
| Compound C | ||
| 0 | −8.13 | 7.43 |
| 0.5 | −7.65 | 7.43 |
| 1 | −7.15 | 7.09 |
| Compound D | ||
| 0 | −8.05 | 7.93 |
| 0.5 | −7.65 | 7.52 |
| 1 | −7.22 | 7.32 |
| Compound | % of Replacement |
|---|---|
| A | 5.70% |
| B | 11.82% |
| C | 9.84% |
| D | 11.33% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marciniak, A.; Krzyżak, E.; Szkatuła, D.; Mazurkiewicz, K.; Kotynia, A. A Spectroscopic and In Silico Description of the Non-Covalent Interactions of Phthalic Acid Imide Derivatives with Deoxyribonucleic Acid—Insights into Their Binding Characteristics and Potential Applications. Molecules 2024, 29, 5422. https://doi.org/10.3390/molecules29225422
Marciniak A, Krzyżak E, Szkatuła D, Mazurkiewicz K, Kotynia A. A Spectroscopic and In Silico Description of the Non-Covalent Interactions of Phthalic Acid Imide Derivatives with Deoxyribonucleic Acid—Insights into Their Binding Characteristics and Potential Applications. Molecules. 2024; 29(22):5422. https://doi.org/10.3390/molecules29225422
Chicago/Turabian StyleMarciniak, Aleksandra, Edward Krzyżak, Dominika Szkatuła, Krystian Mazurkiewicz, and Aleksandra Kotynia. 2024. "A Spectroscopic and In Silico Description of the Non-Covalent Interactions of Phthalic Acid Imide Derivatives with Deoxyribonucleic Acid—Insights into Their Binding Characteristics and Potential Applications" Molecules 29, no. 22: 5422. https://doi.org/10.3390/molecules29225422
APA StyleMarciniak, A., Krzyżak, E., Szkatuła, D., Mazurkiewicz, K., & Kotynia, A. (2024). A Spectroscopic and In Silico Description of the Non-Covalent Interactions of Phthalic Acid Imide Derivatives with Deoxyribonucleic Acid—Insights into Their Binding Characteristics and Potential Applications. Molecules, 29(22), 5422. https://doi.org/10.3390/molecules29225422






