Optimizing Trilobatin Production via Screening and Modification of Glycosyltransferases
Abstract
:1. Introduction
2. Results and Discussion
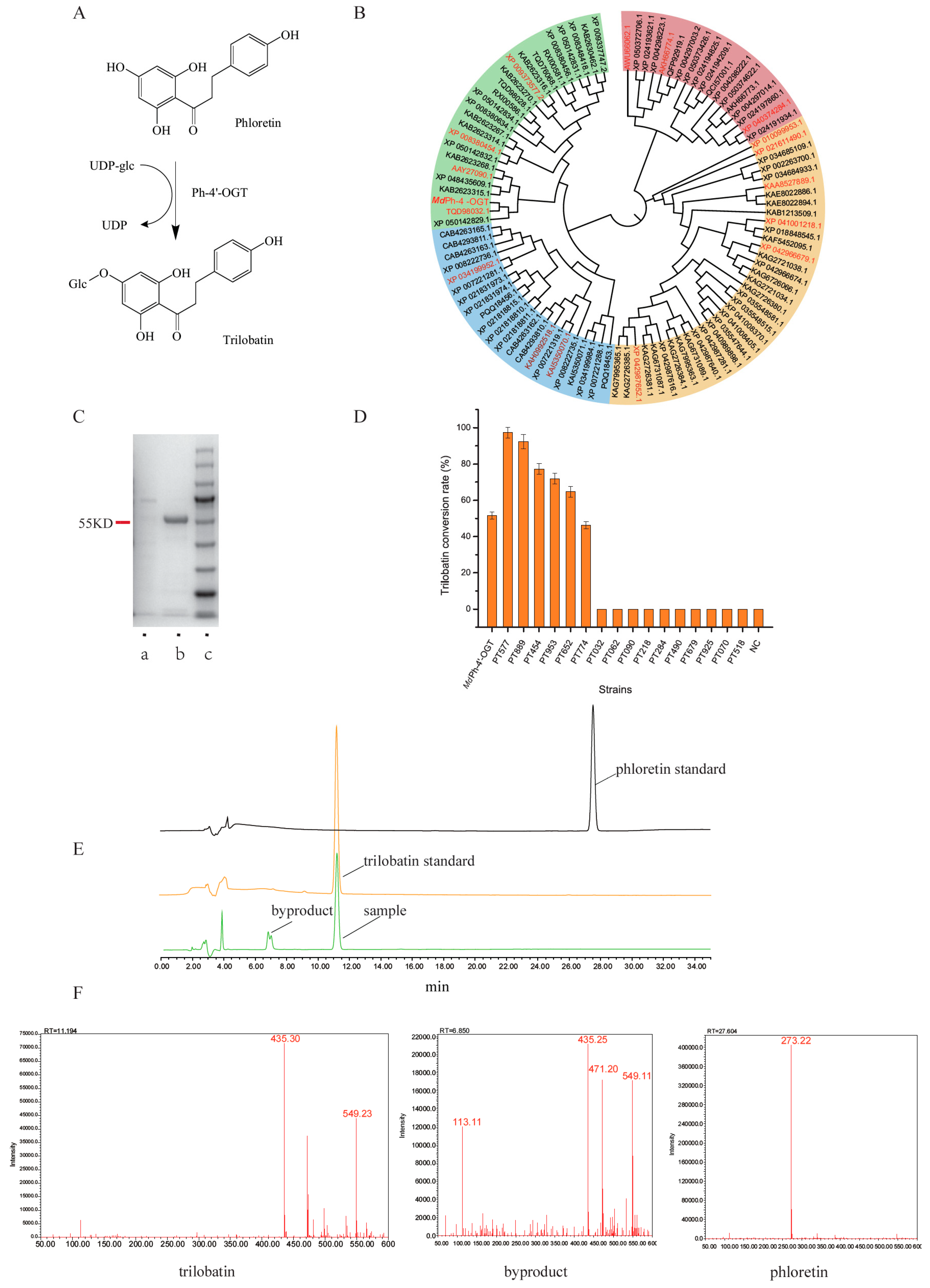
2.1. Screening of ph-4′-OGT
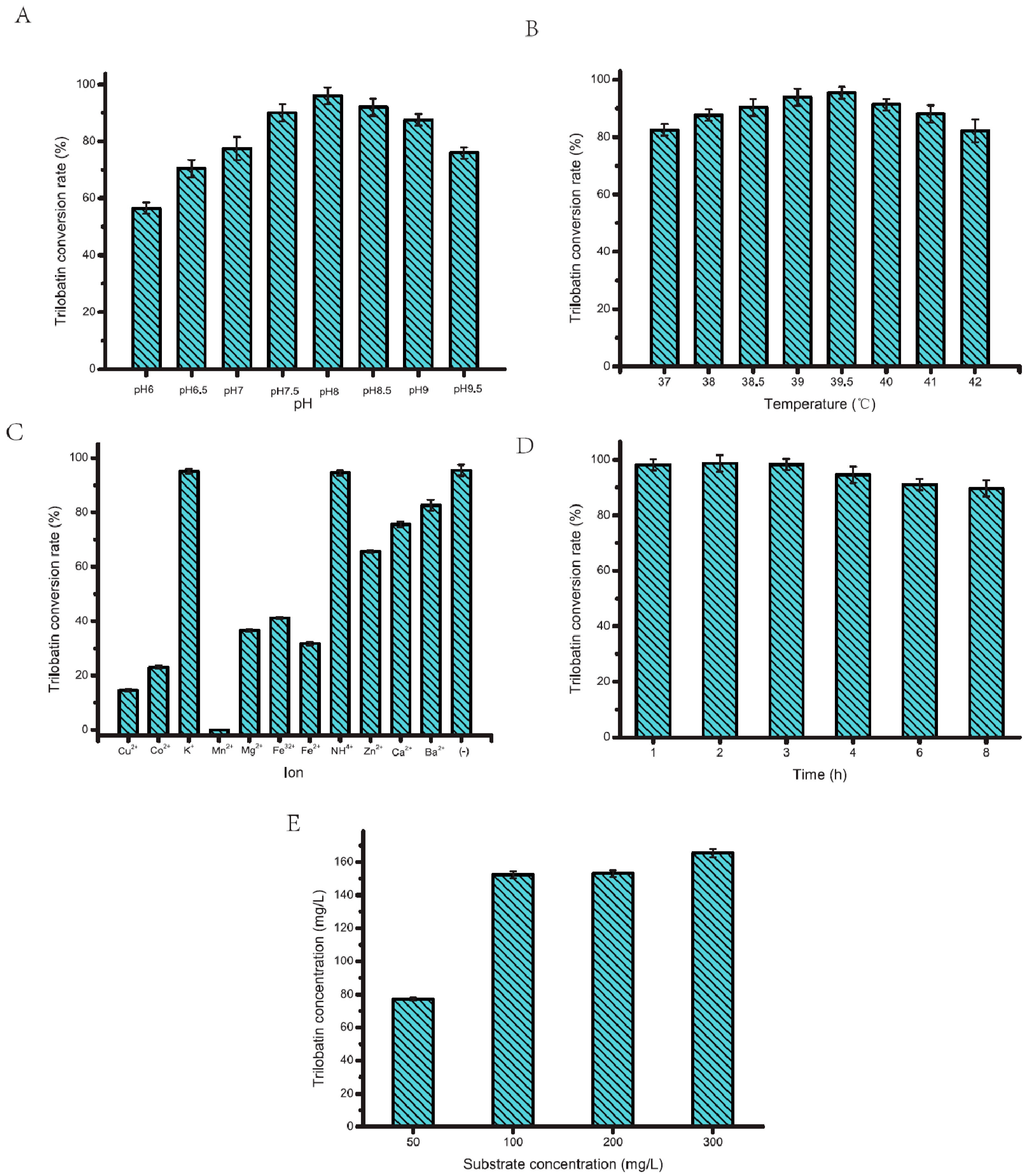
2.2. Optimization of PT577 Glycosyltransferase
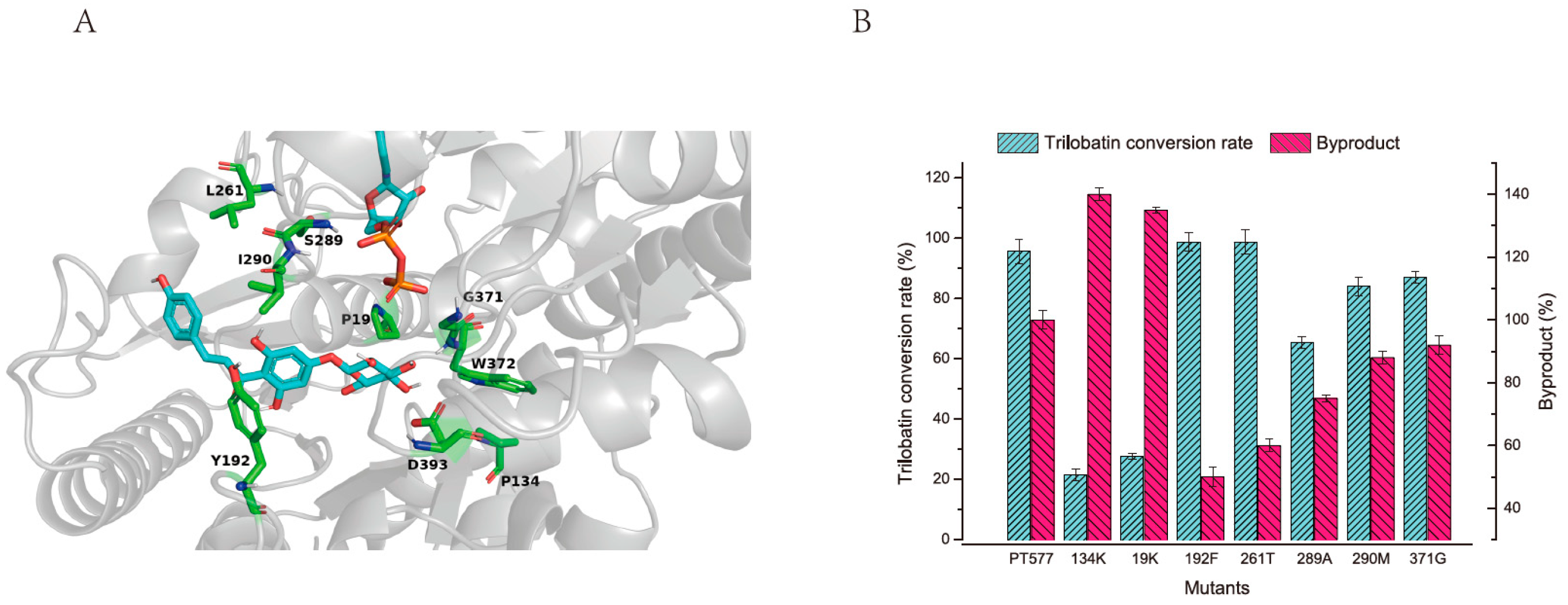
2.3. Modification to Enhance Activity and Specificity of PT577
3. Materials and Methods
3.1. Materials
3.2. Construction of Recombinant Plasmid and Ph-4′-OGT Gene Mining
3.3. Expression of ph-4′-OGT
3.4. Protein Purification of ph-4′-OGT Enzyme
3.5. Enzymatic Reaction with Ph-4′-OGT Enzyme
3.6. Detection of Reaction Products
3.7. Optimization of PT577 Glycosyltransferase
3.8. Modification of PT577
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, J.; Huang, Y.; Yang, Z.; Zhou, C.; Hu, X. Identification and quantitative evaluation of major sweet ingredients in sweet tea (Lithocarpus polystachyus Rehd.) based upon location, harvesting time, leaf age. J. Chem. Soc. Pak. 2018, 40, 158. [Google Scholar]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Després, J.-P.; Hu, F.B. Sugar-sweetened beverages, obesity, type 2 diabetes mellitus, and cardiovascular disease risk. Circulation 2010, 121, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.A.; Sievenpiper, J.L. Controversies about sugars: Results from systematic reviews and meta-analyses on obesity, cardiometabolic disease and diabetes. Eur. J. Nutr. 2016, 55, 25–43. [Google Scholar] [CrossRef]
- Daher, M.; Fahd, C.; Nour, A.A.; Sacre, Y. Trends and amounts of consumption of low-calorie sweeteners: A cross-sectional study. Clin. Nutr. 2022, 48, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Liu, H.-Y.; Luo, M.; Xia, Y.; Yang, X.; Li, H.-Y.; Wu, D.-T.; Sun, Q.; Geng, F.; Gan, R.-Y. Sweet tea (Lithocarpus polystachyus rehd.) as a new natural source of bioactive dihydrochalcones with multiple health benefits. Crit. Rev. Food Sci. 2022, 62, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.; Mu, X.; Liu, J.; Xun, M.; Hu, Y. Study on the differences of metabolites and their bioactivities of Lithocarpus under different processing methods. Food Biosci. 2023, 54, 102817. [Google Scholar] [CrossRef]
- Wang, Y.-K.; Hu, S.-Y.; Xiao, F.-Y.; Dong, Z.-B.; Ye, J.-H.; Zheng, X.-Q.; Liang, Y.-R.; Lu, J.-L. Dihydrochalcones in Sweet Tea: Biosynthesis, Distribution and Neuroprotection Function. Molecules 2022, 27, 8794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiao, M.; Cheng, W.; Song, X.; Wang, S.; Zhao, X.; Dong, J.; Zhang, X.; Long, Y.; Xing, Z. Identification and functional analysis of glycosyltransferase catalyzing the synthesis of phlorizin and trilobatin in Lithocarpus polystachyus Rehd. Ind. Crop. Prod. 2023, 192, 116056. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.-Y.; Xia, Y.; Guo, H.; He, X.-Q.; Li, H.; Wu, D.-T.; Geng, F.; Lin, F.-J.; Li, H.-B. Screening and process optimization of ultrasound-assisted extraction of main antioxidants from sweet tea (Lithocarpus litseifolius [Hance] Chun). Food Biosci. 2021, 43, 101277. [Google Scholar] [CrossRef]
- Shang, A.; Luo, M.; Gan, R.-Y.; Xu, X.-Y.; Xia, Y.; Guo, H.; Liu, Y.; Li, H.-B. Effects of microwave-assisted extraction conditions on antioxidant capacity of sweet tea (Lithocarpus polystachyus Rehd.). Antioxidants 2020, 9, 678. [Google Scholar] [CrossRef]
- Yuan, P.; Xu, M.; Mao, C.; Zheng, H.; Sun, D. Dynamically Regulating Glucose Uptake to Reduce Overflow Metabolism with a Quorum-Sensing Circuit for the Efficient Synthesis of d-Pantothenic Acid in Bacillus subtilis. ACS Synth. Biol. 2023, 12, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Shirai, Y.; Kusubayashi, N.; Karim, M.I.A.; Nakanishi, K.; Hashimoto, K. Effect of organic acid profiles during anaerobic treatment of palm oil mill effluent on the production of polyhydroxyalkanoates by Rhodobacter sphaeroides. J. Ferment. Bioeng. 1996, 82, 151–156. [Google Scholar] [CrossRef]
- van Niel, E.W. Biological processes for hydrogen production. Anaerobes Biotech. 2016, 156, 155–193. [Google Scholar]
- Eichenberger, M.; Lehka, B.J.; Folly, C.; Fischer, D.; Martens, S.; Simón, E.; Naesby, M. Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab. Eng. 2017, 39, 80–89. [Google Scholar] [CrossRef]
- Yahyaa, M.; Davidovich-Rikanati, R.; Eyal, Y.; Sheachter, A.; Marzouk, S.; Lewinsohn, E.; Ibdah, M. Identification and characterization of UDP-glucose: Phloretin 4′-O-glycosyltransferase from Malus x domestica Borkh. Phytochemistry 2016, 130, 47–55. [Google Scholar] [CrossRef]
- Wang, Y.; Yauk, Y.-K.; Zhao, Q.; Hamiaux, C.; Xiao, Z.; Gunaseelan, K.; Zhang, L.; Tomes, S.; López-Girona, E.; Cooney, J. Biosynthesis of the dihydrochalcone sweetener trilobatin requires phloretin glycosyltransferase2. Plant Physiol. 2020, 184, 738–752. [Google Scholar] [CrossRef]
- Gutmann, A.; Bungaruang, L.; Weber, H.; Leypold, M.; Breinbauer, R.; Nidetzky, B. Towards the synthesis of glycosylated dihydrochalcone natural products using glycosyltransferase-catalysed cascade reactions. Green Chem. 2014, 16, 4417–4425. [Google Scholar] [CrossRef]
- Li, H.; Lyv, Y.; Zhou, S.; Yu, S.; Zhou, J. Microbial cell factories for the production of flavonoids-barriers and opportunities. Bioresour. Technol. 2022, 360, 127538. [Google Scholar] [CrossRef]
- Jiang, C.; Liu, X.; Chen, X.; Cai, Y.; Zhuang, Y.; Liu, T.; Zhu, X.; Wang, H.; Liu, Y.; Jiang, H. Raising the production of phloretin by alleviation of by-product of chalcone synthase in the engineered yeast. Sci. China Life Sci. 2020, 63, 1734–1743. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Gibson, D.G. Enzymatic assembly of overlapping DNA fragments. Method Enzymol. 2011, 498, 349–361. [Google Scholar]
- Gibson, D. One-Step Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases in Size. 2009. Available online: https://protocolexchange.researchsquare.com/article/nprot-554/v1 (accessed on 2 December 2023).
- Zhou, K.; Hu, L.; Li, P.; Gong, X.; Ma, F. Genome-wide identification of glycosyltransferases converting phloretin to phloridzin in Malus species. Plant Sci. 2017, 265, 131–145. [Google Scholar] [CrossRef]
- Boyd, S.A.; Mortland, M.M. Enzyme interactions with clays and clay-organic matter complexes. In Soil Biochemistry; Routledge: London, UK, 2017; pp. 1–28. [Google Scholar]
- Van Dyk, J.; Pletschke, B. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes—Factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 2012, 30, 1458–1480. [Google Scholar] [CrossRef]
- Daniel, R.M.; Danson, M.J. A new understanding of how temperature affects the catalytic activity of enzymes. Trends Biochem. Sci. 2010, 35, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R.M.; Danson, M.J. Temperature and the catalytic activity of enzymes: A fresh understanding. FEBS Lett. 2013, 587, 2738–2743. [Google Scholar] [CrossRef]
- Andreini, C.; Bertini, I.; Cavallaro, G.; Holliday, G.L.; Thornton, J.M. Metal ions in biological catalysis: From enzyme databases to general principles. J. Biol. Inorg. Chem. 2008, 13, 1205–1218. [Google Scholar] [CrossRef]
- Lehninger, A. Role of metal ions in enzyme systems. Physiol. Rev. 1950, 30, 393–429. [Google Scholar] [CrossRef] [PubMed]
- Duggleby, R.G. Quantitative analysis of the time courses of enzyme-catalyzed reactions. Methods 2001, 24, 168–174. [Google Scholar] [CrossRef]
- Cao, W.; De La Cruz, E.M. Quantitative full time course analysis of nonlinear enzyme cycling kinetics. Sci. Rep. 2013, 3, 2658. [Google Scholar] [CrossRef] [PubMed]
- German, D.P.; Chacon, S.S.; Allison, S.D. Substrate concentration and enzyme allocation can affect rates of microbial decomposition. Ecology 2011, 92, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Härdin, H.M.; Zagaris, A.; Krab, K.; Westerhoff, H.V. Simplified yet highly accurate enzyme kinetics for cases of low substrate concentrations. FEBS J. 2009, 276, 5491–5506. [Google Scholar] [CrossRef]
- Beuming, T.; Martín, H.; Díaz-Rovira, A.M.; Díaz, L.; Guallar, V.; Ray, S.S. Are deep learning structural models sufficiently accurate for free-energy calculations? Application of FEP+ to AlphaFold2-predicted structures. J. Chem. Inf. Model. 2022, 62, 4351–4360. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Li, Q.; He, W.; Li, M.; Xue, J.; Li, X.; Li, Y. Enhanced biodegradation of modified fluoroquinolone for aerobic, facultative, and anaerobic processes using quantitative structure-activity relationship, molecular docking, and molecular dynamics. Biochem. Eng. J. 2021, 169, 107981. [Google Scholar] [CrossRef]
- Naqvi, S.T.R.; Rasheed, T.; Hussain, D.; ul Haq, M.N.; Majeed, S.; Ahmed, N.; Nawaz, R. Modification strategies for improving the solubility/dispersion of carbon nanotubes. J. Mol. Liq. 2020, 297, 111919. [Google Scholar] [CrossRef]
- Richard, J.P. Protein flexibility and stiffness enable efficient enzymatic catalysis. J. Am. Chem. Soc. 2019, 141, 3320–3331. [Google Scholar] [CrossRef] [PubMed]
- Gulsevin, A.; Meiler, J. An Investigation of Three-Finger Toxin—nAChR Interactions through Rosetta Protein Docking. Toxins 2020, 12, 598. [Google Scholar] [CrossRef] [PubMed]
- Tivon, B.; Gabizon, R.; Somsen, B.A.; Cossar, P.J.; Ottmann, C.; London, N. Covalent flexible peptide docking in Rosetta. Chem. Sci. 2021, 12, 10836–10847. [Google Scholar] [CrossRef]
- Fouret, J.; Brunet, F.G.; Binet, M.; Aurine, N.; Enchéry, F.; Croze, S.; Guinier, M.; Goumaidi, A.; Preininger, D.; Volff, J.-N. Sequencing the genome of Indian flying fox, natural reservoir of nipah virus, using hybrid assembly and conservative secondary scaffolding. Front. Microbiol. 2020, 11, 1807. [Google Scholar] [CrossRef]
- Zeng, L.; Kortschak, R.D.; Raison, J.M.; Bertozzi, T.; Adelson, D.L. Superior ab initio identification, annotation and characterisation of TEs and segmental duplications from genome assemblies. PLoS ONE 2018, 13, e0193588. [Google Scholar] [CrossRef]
- Raissi, V.; Etemadi, S.; Sohrabi, N.; Raiesi, O.; Shahraki, M.; Salimi-Khorashad, A.; Ibrahim, A. Molecular characterization and phylogeny of Taenia hydatigena and Echinococcus granulosus from Iranian sheep and cattle based on COX1 gene. Curr. Microbiol. 2021, 78, 1202–1207. [Google Scholar] [CrossRef]
- Rizwan, M.; Li, Z.; Nie, H.; Qasim, M.; Raza, M.; Hassanyar, A.; Tayyab, M.; Su, S. High mitochondrial diversity of Apis mellifera under COI gene from China and Pakistan. Appl. Ecol. Environ. Res. 2018, 16, 2933–2945. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B.; Shrestha, B.; Bernardini, M.; Hou, J.; Lea, J. DISTEVAL: A web server for evaluating predicted protein distances. BMC Bioinform. 2021, 22, 8. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Cheng, Y.; Bai, T.; Liu, S.; Du, Q.; Xia, W.; Liu, Y.; Wang, X.; Chen, X. Optimizing Trilobatin Production via Screening and Modification of Glycosyltransferases. Molecules 2024, 29, 643. https://doi.org/10.3390/molecules29030643
Yang Y, Cheng Y, Bai T, Liu S, Du Q, Xia W, Liu Y, Wang X, Chen X. Optimizing Trilobatin Production via Screening and Modification of Glycosyltransferases. Molecules. 2024; 29(3):643. https://doi.org/10.3390/molecules29030643
Chicago/Turabian StyleYang, Yue, Yuhan Cheng, Tao Bai, Shimeng Liu, Qiuhui Du, Wenhao Xia, Yi Liu, Xiao Wang, and Xianqing Chen. 2024. "Optimizing Trilobatin Production via Screening and Modification of Glycosyltransferases" Molecules 29, no. 3: 643. https://doi.org/10.3390/molecules29030643




