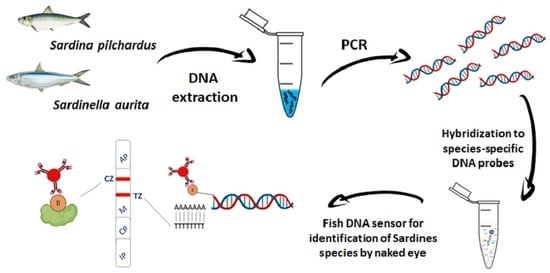
Fish DNA Sensors for Authenticity Assessment—Application to Sardine Species Identification
Abstract
:1. Introduction
2. Results
2.1. Effect of the Treatment of the Sample to the Optical Signal of the Test Zone of the Strip
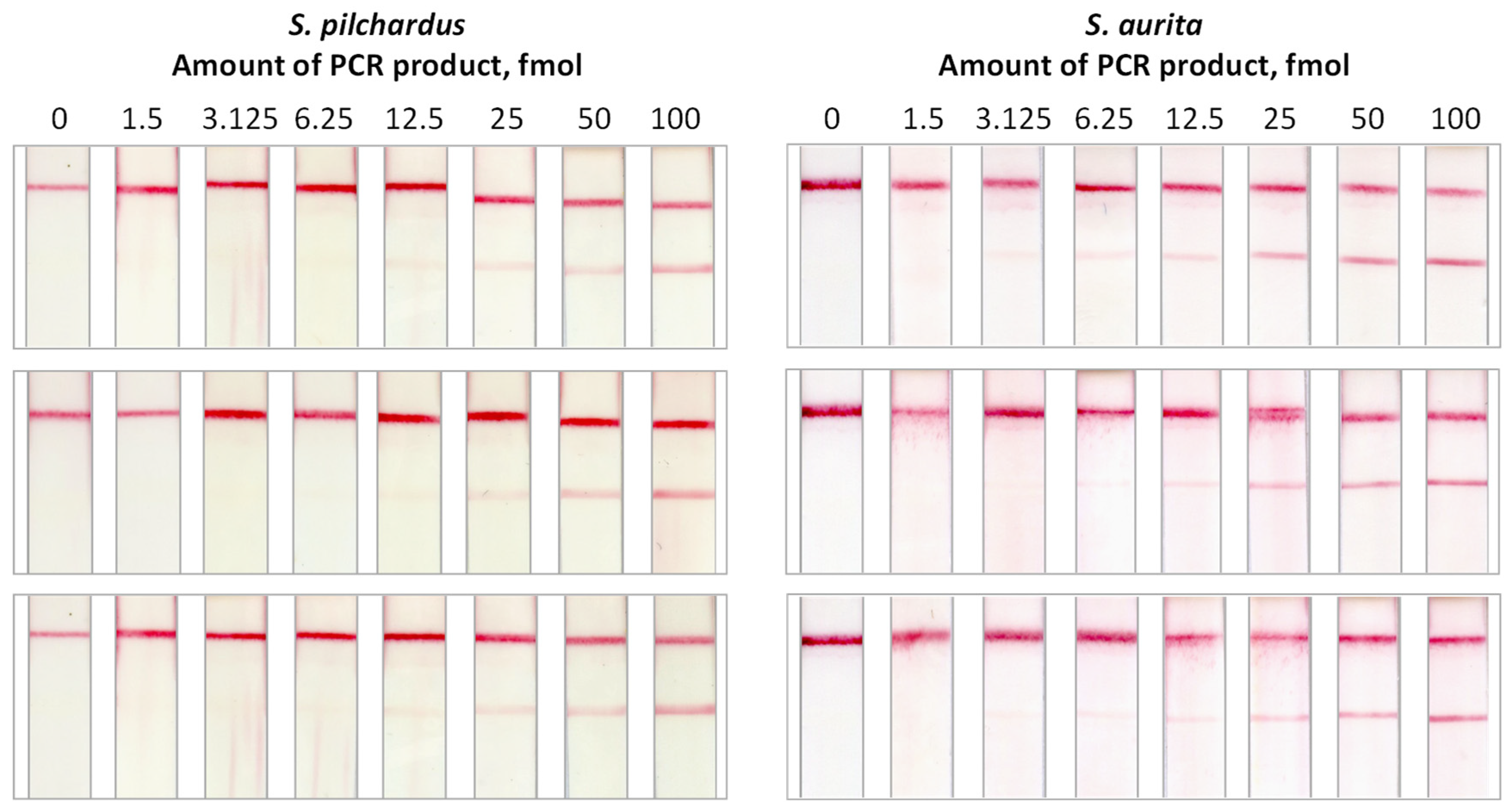
2.2. Effect of the Amount of the Amplification Product on the Signal
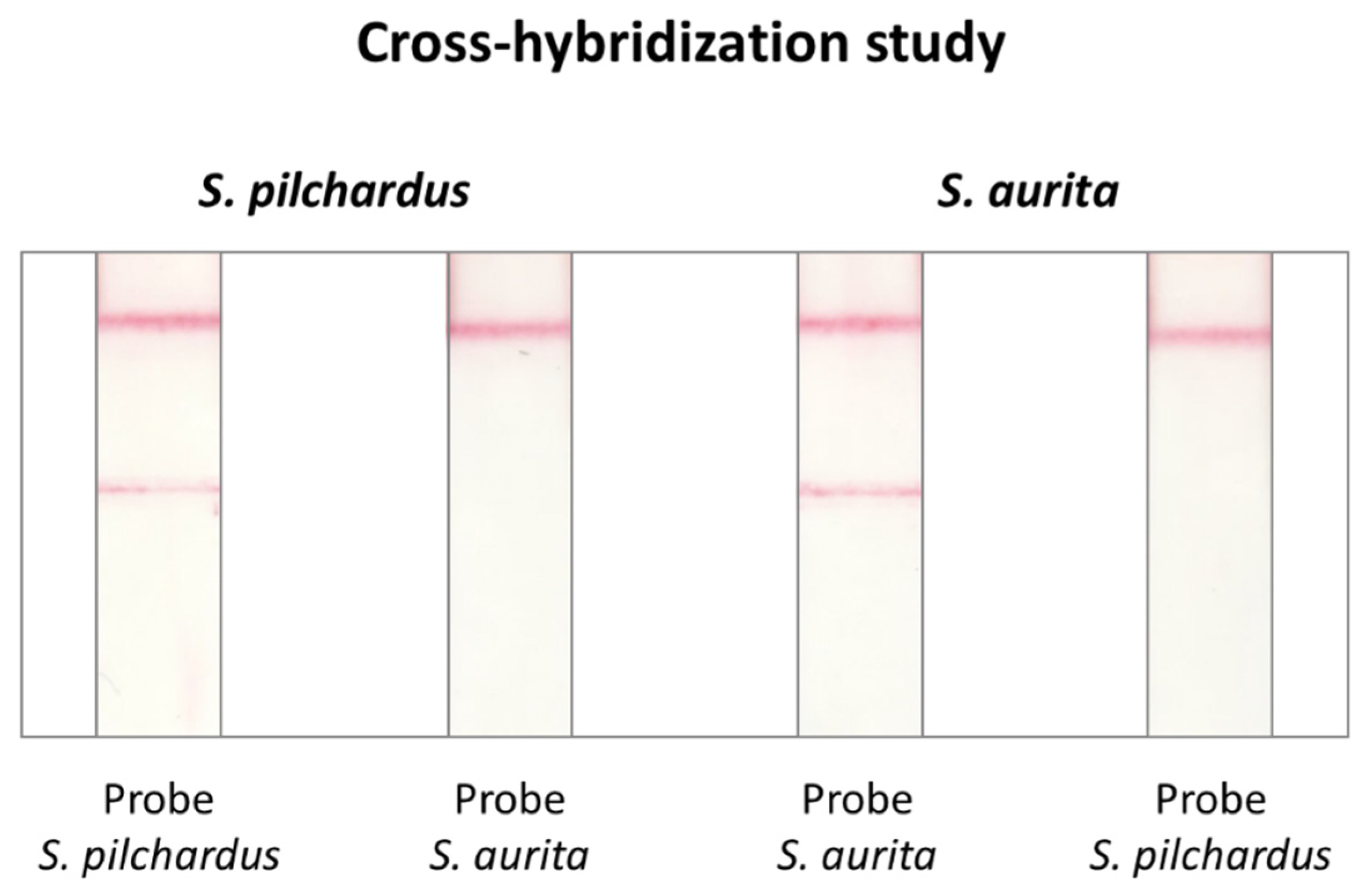
2.3. Cross-Hybridization Study
2.4. Method Performance Evaluation Using Mixtures of Amplification Products
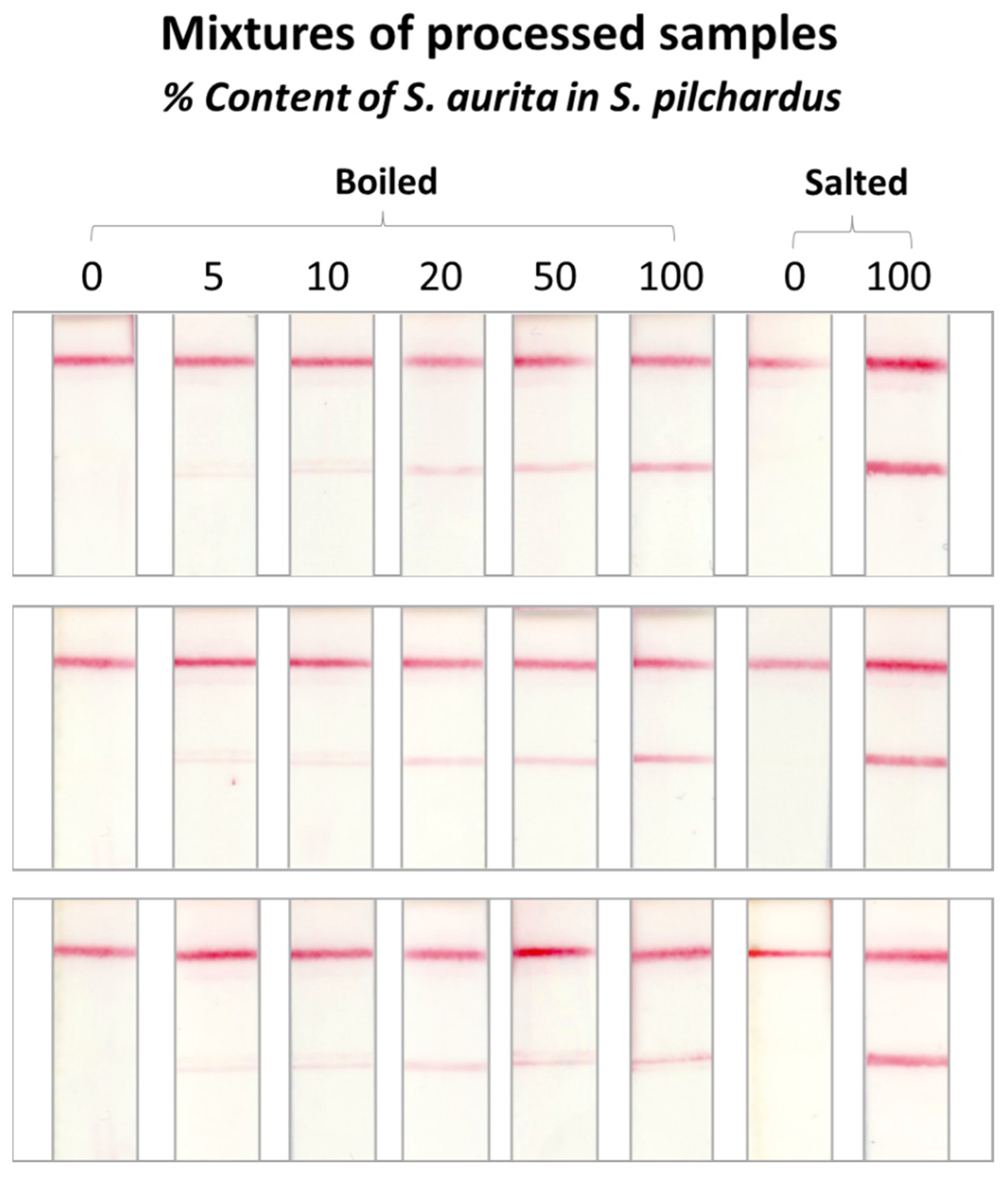
2.5. Method Performance Evaluation Using Mixtures of Processed Samples
2.6. Repeatability of the DNA Sensor
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Samples
4.3. Sample Preparation
4.4. DNA Extraction
4.5. Primer Design
4.6. Polymerase Chain Reaction (PCR)
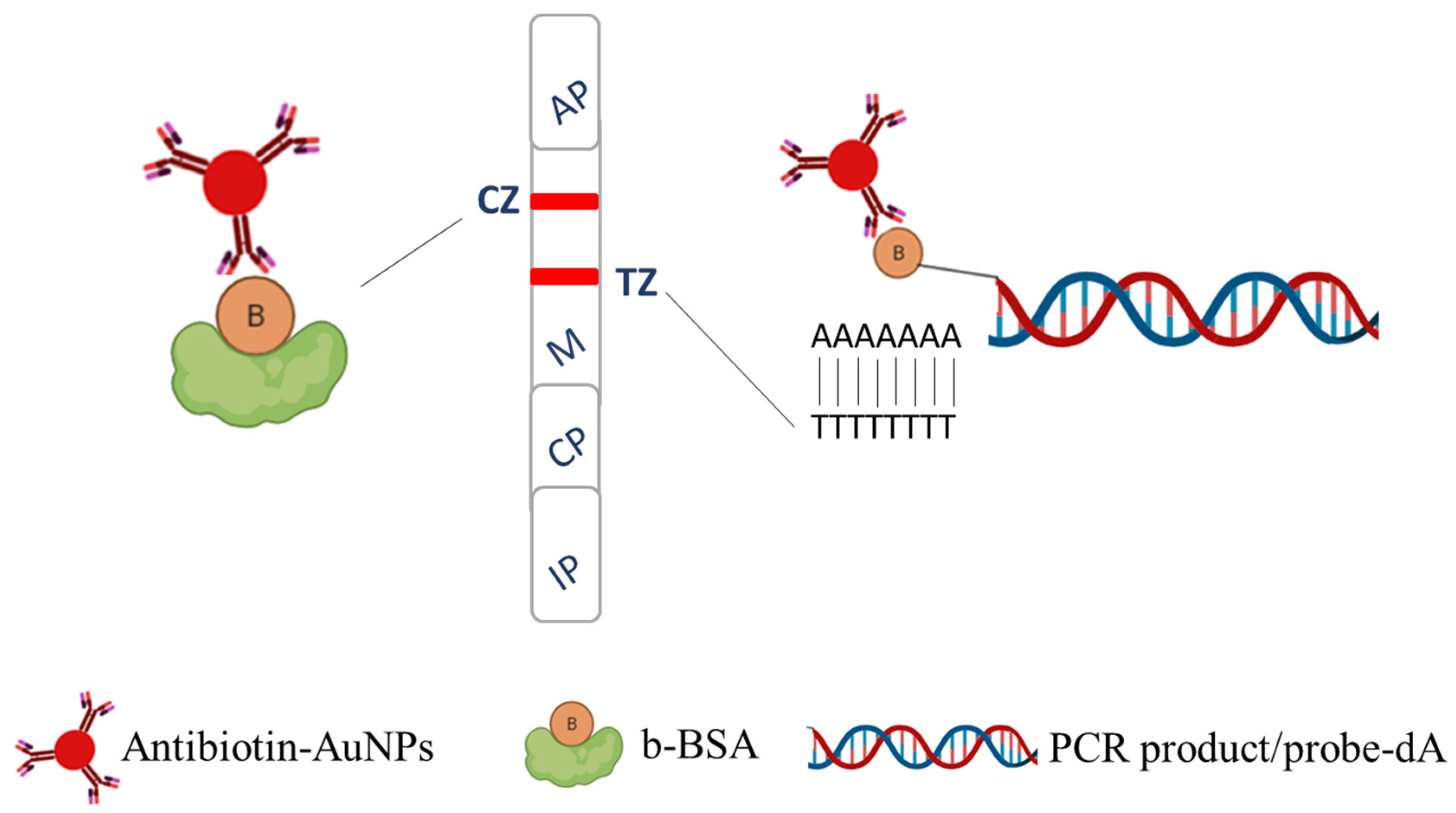
4.7. Construction of the Sensing Zones of the Device
4.7.1. Biotinylation of Bovine Serum Albumin (b-BSA)
4.7.2. Insertion of a Poly-dT Tail into the Capture Probe
4.8. Insertion of a Poly-dA Tail into the Detection Probes
4.9. Coupling of Anti-Biotin Antibody to Gold Nanoparticles (Antibiotin-AuNPs)
4.10. Assembly of the Fish DNA Sensor
4.11. Detection of S. pilchardus and S. aurita
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smaoui, S.; Tarapoulouzi, M.; Agriopoulou, S.; D’Amore, T.; Varzakas, T. Current state of milk, dairy products, meat and meat products, eggs, fish and fishery products authentication and chemometrics. Foods 2023, 12, 4254. [Google Scholar] [CrossRef] [PubMed]
- Kotsanopoulos, K.V.; Exadactylos, A.; Gkafas, G.A.; Martsikalis, P.V.; Parlapani, F.F.; Boziaris, I.S.; Arvanitoyannis, I.S. The use of molecular markers in the verification of fish and seafood authenticity and the detection of adulteration. Compr. Rev. Food Sci. Food Saf. 2021, 20, 1584–1654. [Google Scholar] [CrossRef]
- Cermakova, E.; Lencova, S.; Mukherjee, S.; Horka, P.; Vobruba, S.; Demnerova, K.; Zdenkova, K. Identification of fish species and targeted genetic modifications based on DNA analysis: State of the Art. Foods 2023, 12, 228. [Google Scholar] [CrossRef]
- Chaudhary, V.; Kajla, P.; Dewan, A.; Pandiselvam, R.; Socol, C.T.; Maerescu, C.M. Spectroscopic techniques for authentication of animal origin foods. Front. Nutr. 2022, 9, 979205. [Google Scholar] [CrossRef]
- Herrero, B.; Lago, F.C.; Vieites, J.M.; Espiñeira, M. Development of a rapid and simple molecular identification methodology for true sardines (Sardina pilchardus) and false sardines (Sardinella aurita) based on the real-time PCR technique. Eur. Food Res. Technol. 2011, 233, 851–857. [Google Scholar] [CrossRef]
- Lago, F.C.; Herrero, B.; Vieites, J.M.; Espiñeira, M. FINS Methodology to identification of sardines and related species in canned products and detection of mixture by means of SNP analysis systems. Eur. Food Res. Technol. 2011, 232, 1077–1086. [Google Scholar] [CrossRef]
- Dawan, S.J.; Ahn, J. Application of DNA barcoding for ensuring food safety and quality. Food Sci. Biotechnol. 2022, 31, 1355–1364. [Google Scholar] [CrossRef]
- Zaukuu, J.L.Z.; Benes, E.; Bázár, G.; Kovács, Z.; Fodor, M. Agricultural potentials of molecular spectroscopy and advances for food authentication: An overview. Processes 2022, 10, 214. [Google Scholar] [CrossRef]
- Wang, X.Y.; Xie, J.; Chen, X.J. Applications of non-invasive and novel methods of low-field nuclear magnetic resonance and magnetic resonance imaging in aquatic products. Front. Nutr. 2021, 8, 651804. [Google Scholar] [CrossRef]
- Hassoun, A.; Sahar, A.; Lakhal, L.; Aït-Kaddour, A. Fluorescence spectroscopy as a rapid and non-destructive method for monitoring quality and authenticity of fish and meat products: Impact of different preservation conditions. LWT 2019, 103, 279–292. [Google Scholar] [CrossRef]
- Franco, C.M.; Ambrosio, R.L.; Cepeda, A.; Anastasio, A. Fish intended for human consumption: From DNA barcoding to a next-generation sequencing (NGS)-based approach. Curr. Opin. Food Sci. 2021, 42, 86–92. [Google Scholar] [CrossRef]
- Jérôme, M.; Lemaire, C.; Bautista, J.M.; Fleurence, J.; Etienne, M. Molecular phylogeny and species identification of sardines. J. Agric. Food Chem. 2003, 51, 43–50. [Google Scholar] [CrossRef]
- Leonardo, R.; Nunes, R.S.C.; Monteiro, M.L.G.; Conte-Junior, C.A.; Del Aguila, E.M.; Paschoalin, V.M.F. Molecular testing on sardines and rulings on the authenticity and nutritional value of marketed fishes: An experience report in the state of Rio de Janeiro, Brazil. Food Control 2016, 60, 394–400. [Google Scholar] [CrossRef]
- Besbes, N.; Fattouch, S.; Sadok, S. Differential detection of small pelagic fish in tunisian canned products by PCR-RFLP: An efficient tool to control the label information. Food Control 2012, 25, 260–264. [Google Scholar] [CrossRef]
- Jérôme, M.; Lemaire, C.; Verrez-Bagnis, V.; Etienne, M. Direct sequencing method for species identification of canned sardine and sardine-type products. J. Agric. Food Chem. 2003, 51, 7326–7332. [Google Scholar] [CrossRef]
- Labrador, K.; Agmata, A.; Palermo, J.D.; Follante, J.; Pante, M.J. Authentication of processed Philippine sardine products using hotshot DNA extraction and minibarcode amplification. Food Control 2019, 98, 150–155. [Google Scholar] [CrossRef]
- Bottero, M.T.; Dalmasso, A.; Nucera, D.; Turi, R.M.; Rosati, S.; Squadrone, S.; Goria, M.; Civera, T. Development of a PCR assay for the detection of animal tissues in ruminant feeds. J. Food Prot. 2003, 66, 2307–2312. [Google Scholar] [CrossRef] [PubMed]
- Durand, J.D.; Diatta, M.A.; Diop, K.; Trape, S. Multiplex 16S rRNA haplotype-specific PCR, a rapid and convenient method for fish species identification: An application to West African Clupeiform larvae. Mol. Ecol. Resour. 2010, 10, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Besbes, N.; Fattouch, S.; Sadok, S. Comparison of methods in the recovery and amplificability of DNA from fresh and processed sardine and anchovy muscle tissues. Food Chem. 2011, 129, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Atarhouch, T.; Rami, M.; Naciri, M.; Dakkak, A. Genetic population structure of sardine (Sardina pilchardus) off Morocco detected with intron polymorphism (EPIC-PCR). Mar. Biol. 2007, 150, 521–528. [Google Scholar] [CrossRef]
- Xiong, X.; Yuan, F.; Huang, M.; Xiong, X. Exploring the possible reasons for fish fraud in China based on results from monitoring sardine products sold on Chinese markets using DNA barcoding and real time PCR. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2020, 37, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Cuende, E.; Mendibil, I.; Bachiller, E.; Álvarez, P.; Cotano, U.; Rodriguez-Ezpeleta, N. A real-time PCR approach to detect predation on anchovy and sardine early life stages. J. Sea Res. 2017, 130, 204–209. [Google Scholar] [CrossRef]
- Armani, A.; Castigliego, L.; Tinacci, L.; Gianfaldoni, D.; Guidi, A. Multiplex conventional and real-time PCR for fish species identification of Bianchetto (juvenile form of Sardina pilchardus), Rossetto (Aphia minuta), and Icefish in fresh, marinated and cooked products. Food Chem. 2012, 133, 184–192. [Google Scholar] [CrossRef]
- Bréchon, A.L.; Coombs, S.H.; Sims, D.W.; Griffiths, A.M. Development of a rapid genetic technique for the identification of Clupeid larvae in the Western English channel and investigation of mislabelling in processed fish products. ICES J. Mar. Sci. 2013, 70, 399–407. [Google Scholar] [CrossRef]



| Name | Sequence (5′→3′) | |
|---|---|---|
| Primers | Sard Forward | Biotin-CGGTGGTMAAACACATG |
| Sard Reverse | GTCTGATCTGAGGTCGT | |
| Probes | S. pilchardus Probe | CCTTGCTCCAGAGGTCCG |
| S. aurita Probe | CCTYGCTCTACGGTCCGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kakarelidou, M.; Christopoulos, P.; Conides, A.; Kalogianni, D.P.; Christopoulos, T.K. Fish DNA Sensors for Authenticity Assessment—Application to Sardine Species Identification. Molecules 2024, 29, 677. https://doi.org/10.3390/molecules29030677
Kakarelidou M, Christopoulos P, Conides A, Kalogianni DP, Christopoulos TK. Fish DNA Sensors for Authenticity Assessment—Application to Sardine Species Identification. Molecules. 2024; 29(3):677. https://doi.org/10.3390/molecules29030677
Chicago/Turabian StyleKakarelidou, Myrto, Panagiotis Christopoulos, Alexis Conides, Despina P. Kalogianni, and Theodore K. Christopoulos. 2024. "Fish DNA Sensors for Authenticity Assessment—Application to Sardine Species Identification" Molecules 29, no. 3: 677. https://doi.org/10.3390/molecules29030677
APA StyleKakarelidou, M., Christopoulos, P., Conides, A., Kalogianni, D. P., & Christopoulos, T. K. (2024). Fish DNA Sensors for Authenticity Assessment—Application to Sardine Species Identification. Molecules, 29(3), 677. https://doi.org/10.3390/molecules29030677