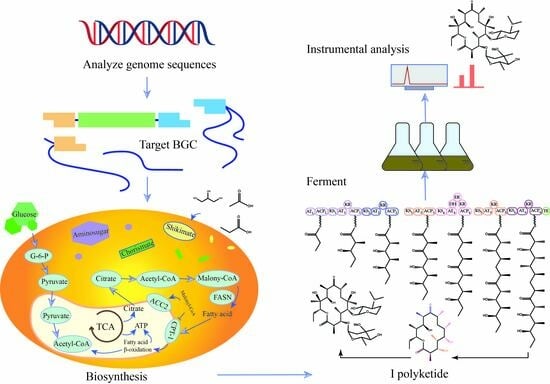
Effect of Precursors and Their Regulators on the Biosynthesis of Antibiotics in Actinomycetes
Abstract
:1. Introduction
2. Secondary Metabolite of Actinomycetes
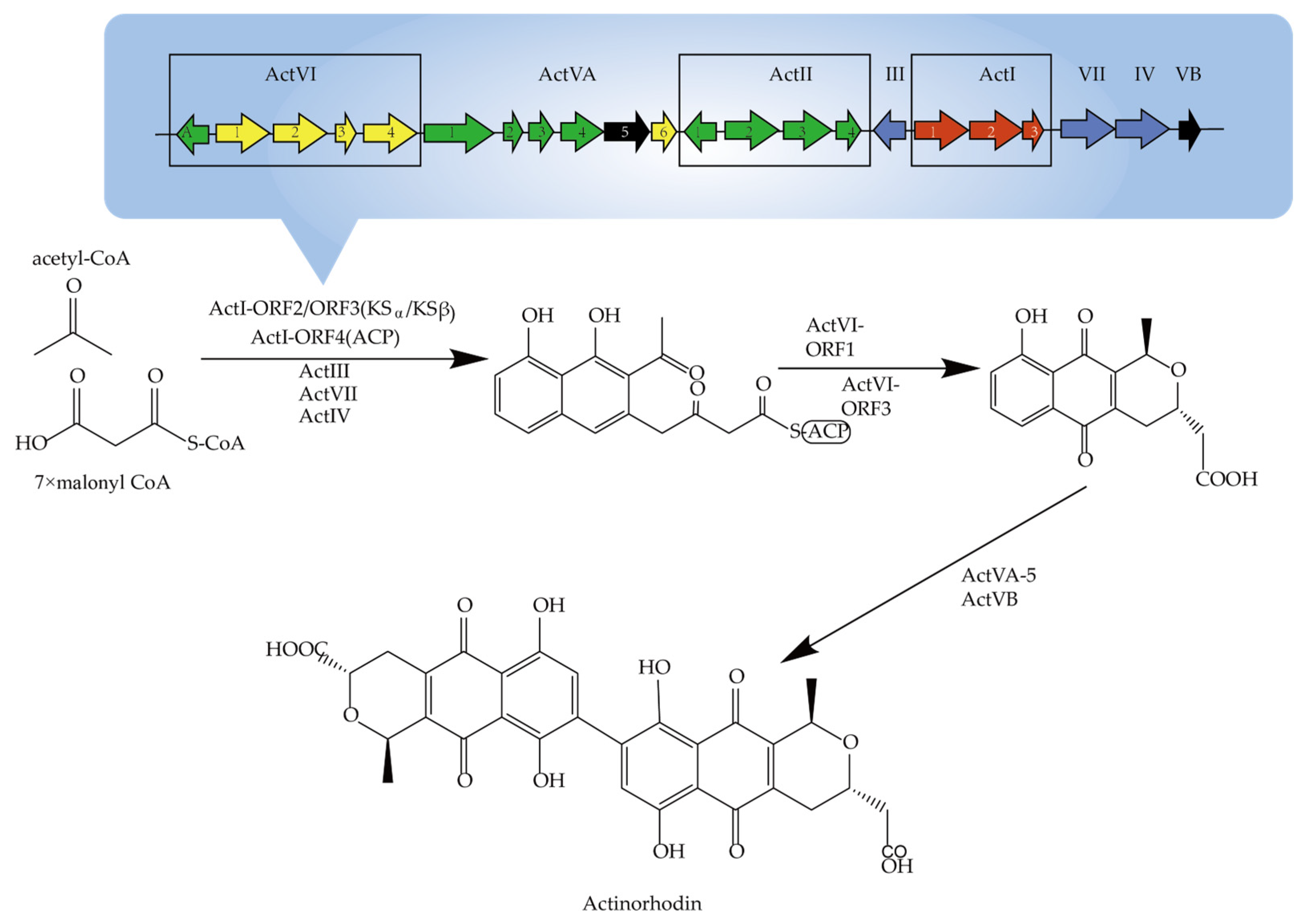
2.1. Secondary Metabolite and Biosynthesis Pathway in Actinomycetes
2.2. Potential Secondary Metabolites of Actinomycetes
3. Regulation of Secondary Metabolite Synthesis in Actinomycetes
3.1. Pathway-Specific Regulation in Secondary Metabolites
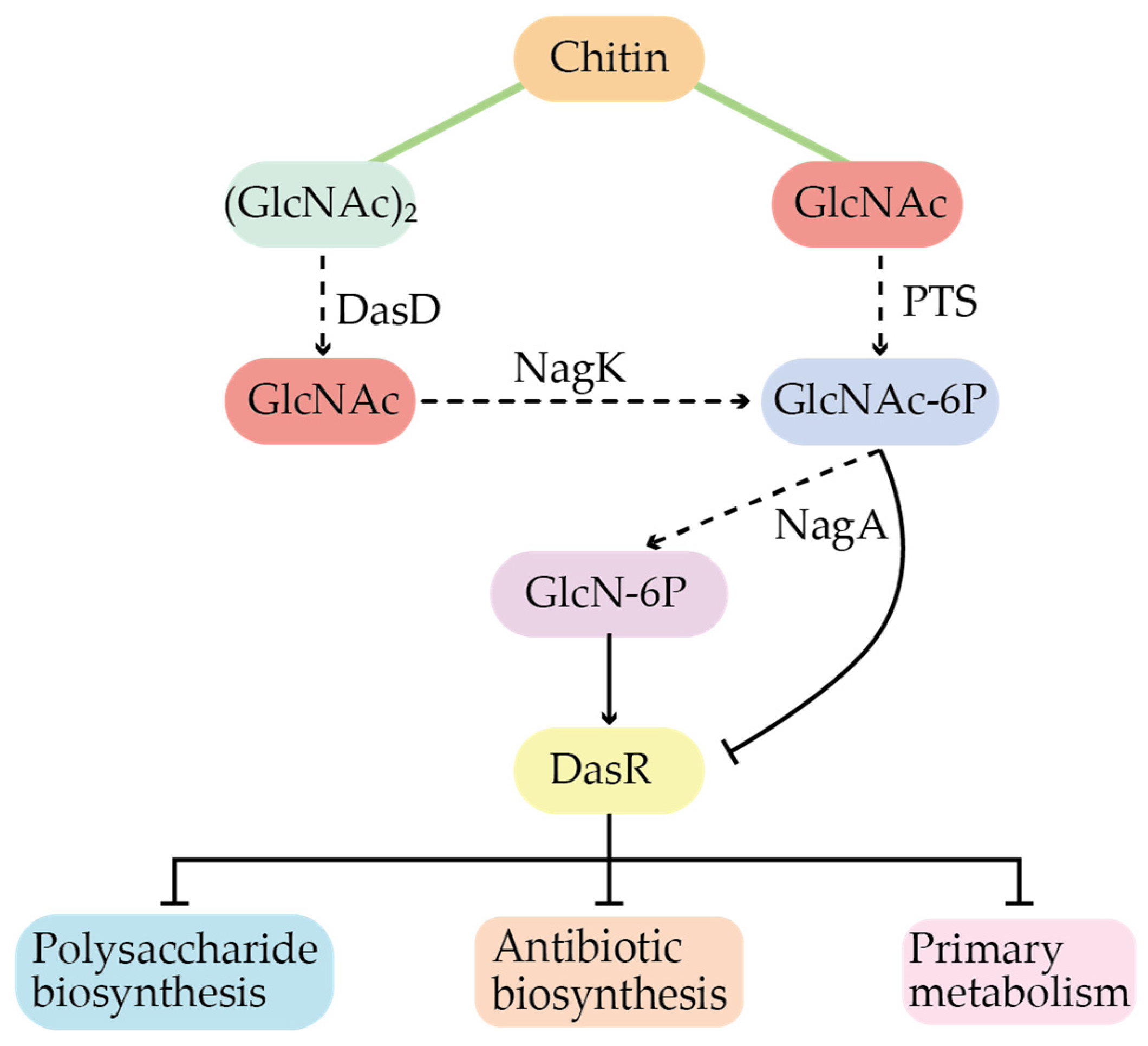
3.2. Global Regulation in Secondary Metabolites
4. Effect of Precursors on Antibiotic Biosynthesis
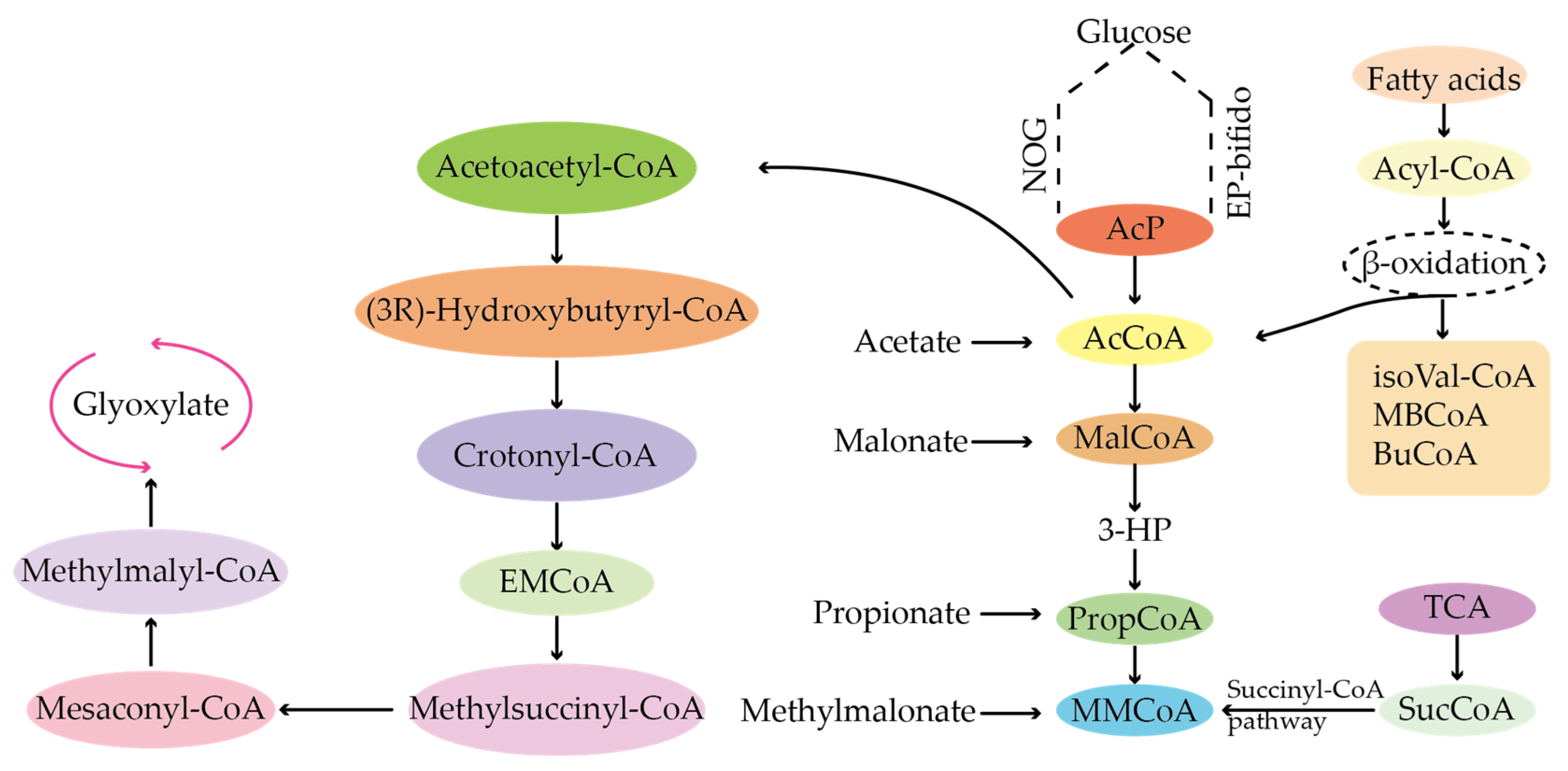
4.1. Types of Precursors of Secondary Metabolites
4.2. Sources of Precursors for the Biosynthesis of Antibiotics
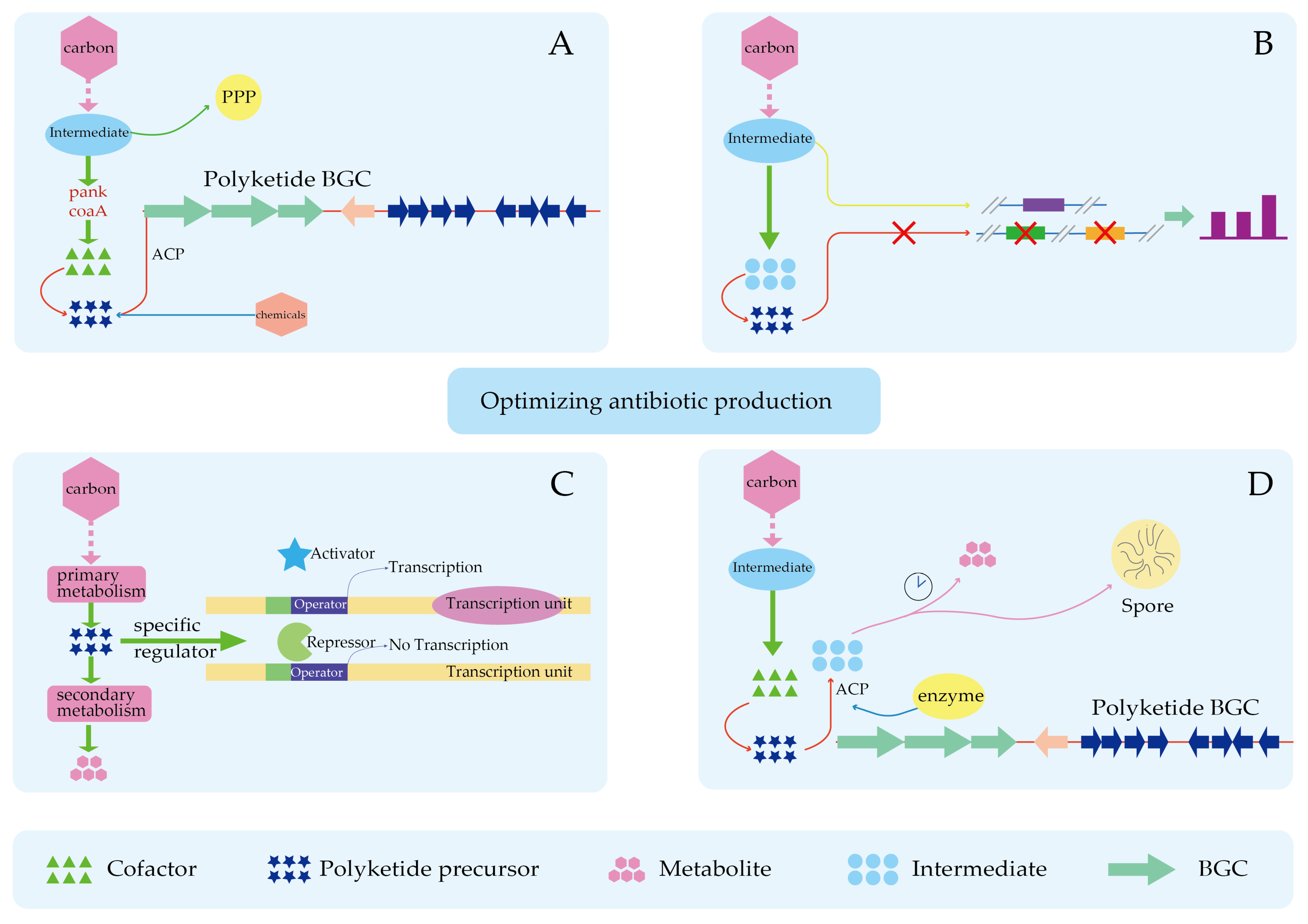
4.3. Improved Precursor Supply Can Partially Relieve the Limitation of Product Synthesis by Insufficient Precursors
5. TetR Family Proteins Regulate Acyl-CoA-like Precursor Synthesis via a Receptor–Ligand Model
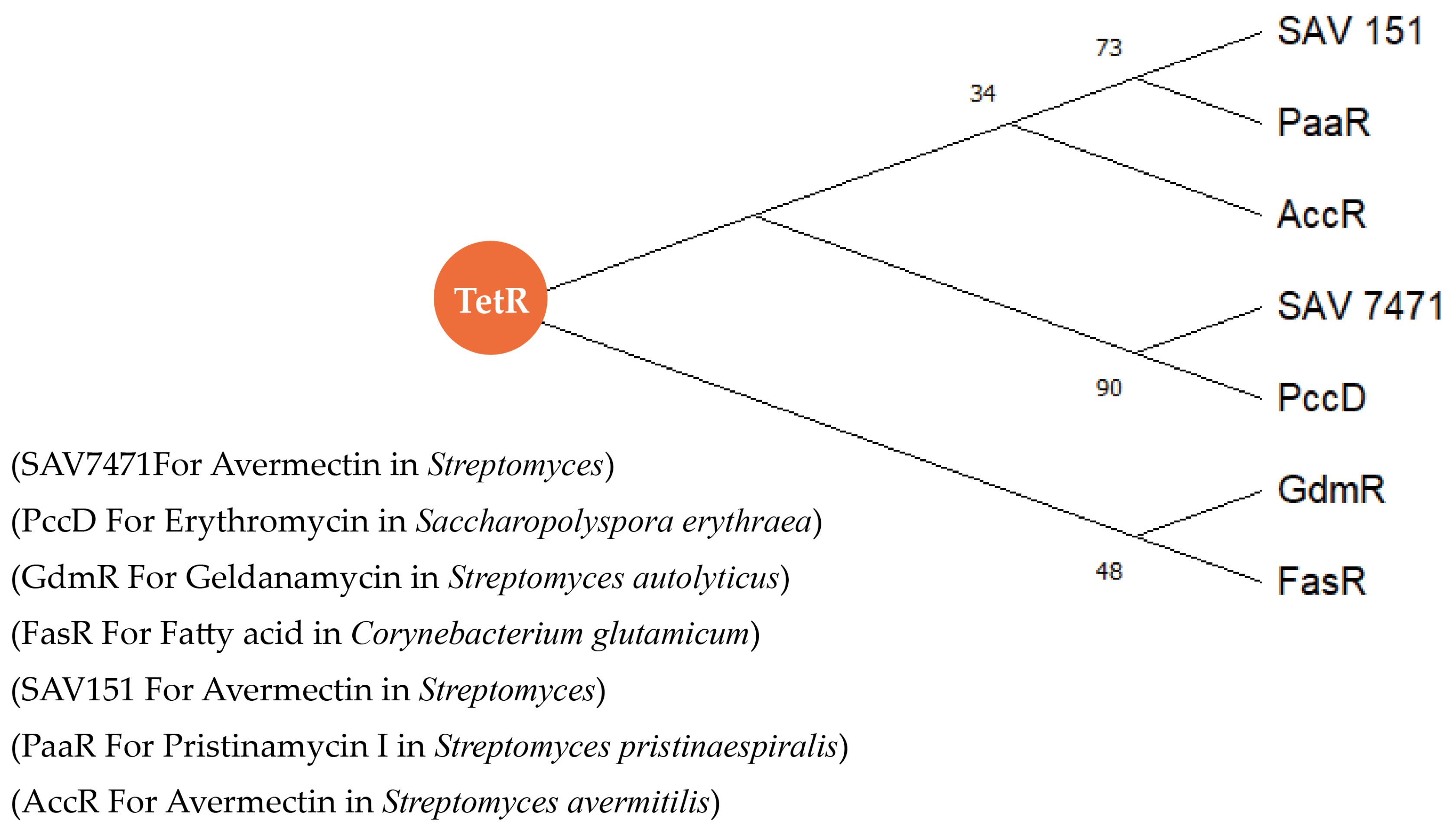
5.1. The TetR Family of Regulators
5.2. Regulation of Product Synthesis by Receptor–Ligand Pattern in the Typical Type I PKS Synthesis Pathway
5.3. TetR Family Regulators Modulate Precursor Synthesis
6. New Approach in Precursor Research
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mervyn, B. Regulation of Secondary Metabolism in Streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar]
- Malpartida, F.; Hopwood, D. Molecular Cloning of the Whole Biosynthetic Pathway of a Streptomyces Antibiotic and its Expression in a Heterologous Host. Nature 1984, 309, 462–464. [Google Scholar] [CrossRef]
- Laureti, L.; Song, L.; Huang, S.; Corre, C.; Leblond, P.; Challis, G.L. Identification of a Bioactive 51-Membered Macrolide Complex by Activation of a Silent Polyketide Synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 2011, 108, 6258–6263. [Google Scholar] [CrossRef] [PubMed]
- Sidda, J.-D.; Song, L.; Poon, V.; Al–Bassam, M.; Lazos, O.; Buttner, M.J.; Challis, G.L.; Corre, C. Discovery of a Family of γ-Aminobutyrate Ureas Via Rational Derepression of a Silent Bacterial Gene Cluster. Chem. Sci. 2014, 5, 86–89. [Google Scholar] [CrossRef]
- Saleh, O.; Bonitz, T.; Flinspach, K.; Kulik, A.; Burkard, N.; Mühlenweg, A.; Vente, A.; Polnick, S.; Lämmerhofer, M.; Gust, B.; et al. Activation of a Silent Phenazine Biosynthetic Gene Cluster Reveals a Novel Natural Product and a New Resistance Mechanism Against Phenazines. Med. Chem. Commun. 2012, 3, 1009–1019. [Google Scholar] [CrossRef]
- Kawachi, R.; Wangchaisoonthorn, U.; Nihira, T.; Yamada, Y. Identification by Gene Deletion Analysis of a Regulator, VmsR, that Controls Virginiamycin Biosynthesis in Streptomyces virginiae. J. Bacteriol. 2000, 11, 6259–6263. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-J.; Shan, Y.-M.; Li, H.; Dou, W.-W.; Jiang, X.-H.; Mao, X.-M.; Liu, S.-P.; Guan, W.-J.; Li, Y.-Q. Multiple transporters are involved in natamycin efflux in Streptomyces chattanoogensis L10. Mol. Microbiol. 2017, 103, 713–728. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.-H.; Zhou, R.-C.; Chen, X.-P.; Luo, S.; Wang, F.; Mao, X.-M.; Li, Y.-Q. DepR1, a TetR Family Transcriptional Regulator, Positively Regulates Daptomycin Production in an Industrial Producer, Streptomyces roseosporus SW0702. Appl. Environ. Microbiol. 2016, 82, 1898–1905. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.-J.; Hwang, J.-Y.; Shin, H.-L.; Cui, H.-Q.; Lee, J.; Yoon, Y.-J. Characterization of Negative Regulatory Genes for the Biosynthesis of Rapamycin in Streptomyces rapamycinicus and its Application for Improved Production. Ind. Microbiol. Biotechnol. 2015, 42, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Wei, W.; Jin, X.; Ge, M.; Chen, D.-J. Influence of Tet R family transcriptional regulatory gene tfpA on A21978C production in Streptomyces. Chin. J. Antibiot. 2015, 5, 334–337, 368. [Google Scholar]
- Bate, N.; Butler, A.-R.; Gandecha, A.-R.; Cundliffe, E. Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem. Biol. 1999, 6, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Rang, J.; Xia, Z.; Shuai, L.; Cao, L.; Liu, Y.; Li, X.-M.; Xie, J.; Li, Y.-L.; Hu, S.-B.; Xie, Q.-J.; et al. A TetR Family Transcriptional Regulator, SP_2854 can Affect the Butenyl-Spinosyn Biosynthesis by Regulating Glucose Metabolism in Saccharopolyspora pogona. Microb. Cell Fact. 2022, 21, 83. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Hong, B. Advances in the Mechanisms of Secondary Metabolism Regulation in Streptomyces. Chin. J. Biotechnol. 2004, 24, 39–42, 47. [Google Scholar]
- Martín-Martín, S.; Rodríguez-García, A.; Santos-Beneit, F.; Franco-Domínguez, E.; Sola-Landa, A.; Martín, F.-J. Self-Control of the PHO Regulon: The PhoP-Dependent Protein PhoU Controls Negatively Expression of Genes of PHO Regulon in Streptomyces coelicolor. J. Antibiot. 2017, 71, 113–122. [Google Scholar] [CrossRef]
- Lee, P.-C.; Umeyama, T.; Horinouchi, S. AfsS is a Target of AfsR, A Transcriptional Factor with ATPase Activity that Globally Controls Secondary Metabolism in Streptomyces coelicolor A3. Mol. Microbiol. 2002, 43, 1413–1430. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-Y.; Guo, W.-W.; Geng, M.-X.; Zhuang, Z.-C.; Bai, L.-P. Global Regulatory Protein DasR in Streptomyces. Acta Microbiol. Sin. 2022, 62, 1260–1269. [Google Scholar]
- Valente, A.-M.; Ferreira, A.-G.; Daolio, C.; Filho, E.-R.; Boffo, E.-F.; Souza, A.-Q.; Sebastianes, F.-L.; Melo, I.-S. Production of 5-Hydroxy-7-Methoxy-4-Methylphthalide in a Culture of Penicillium crustosum. An. Acad. Bras. Cienc. 2013, 85, 487–496. [Google Scholar] [CrossRef]
- Wang, F.-Z.; Wei, H.-J.; Zhu, T.-J.; Li, D.-H.; Lin, Z.-J.; Gu, Q.-Q. Three New Cytochalasins From the Marine-Derived Fungus Spicaria Elegans KLA03 by Supplementing the Cultures with L- and D-tryptophan. Chem. Biodivers. 2011, 8, 887–894. [Google Scholar] [CrossRef]
- Betina, V. Biosynthetic Pathways of Secondary Metabolites. Amsterdam. Online 1994, 5, 66. [Google Scholar]
- Leitão, A.L.; Enguita, F.J. Chapter 4-Synthetic Biology Approaches for Secondary Metabolism Engineering. In Microbial Cell Factories Engineering for Production of Biomolecules; Academic Press: Cambridge, MA, USA, 2021; pp. 51–64. [Google Scholar]
- Chu, J. The Role of Special Precursors in Antibiotic Biosynthesis. World Notes Antibiot. 1999, 20, 202–212. [Google Scholar]
- Elsayed, E.-A.; Farid, M.-A.-F.; Enshasy, H.-A.-E. Improvement in natamycin production by Streptomyces natalensis with the addition of short-chain carboxylic acids. Process Biochem. 2013, 12, 1831–1838. [Google Scholar] [CrossRef]
- Recio, E.; Aparicio, J.-F.; Rumbero, Á.; Martín, J.-F. Glycerol, ethylene glycol and propanediol elicit pimaricin biosynthesis in the PI-factor-defective strain Streptomyces natalensis npi287 and increase polyene production in several wild-type actinomycetes. J. Microbiol. 2006, 152 Pt 10, 3147–3156. [Google Scholar] [CrossRef]
- Mo, S.-J.; Ban, Y.-H.; Park, J.-W.; Yoo, Y.-J.; Yoon, Y.-J. Enhanced FK506 Production in Streptomyces clavuligerus CKD1119 by Engineering the Supply of Methylmalonyl-CoA Precursor. Ind. Microbiol. Biotechnol. 2009, 36, 1473–1482. [Google Scholar] [CrossRef]
- Kong, D.-Z.; LI, H.; LI, X.-J.; Xie, Z.-J.; Liu, H. Engineering the Precursor Supply Pathway in Streptomyces gilvosporeus for Overproduction of Natamycin. Chin. J. Biotechnol. 2022, 38, 4630–4643. [Google Scholar]
- Zhang, J.-Y.; Xu, J.; Li, H.-J.; Zhang, Y.-Y.; Ma, Z.; Bechthold, A.; Yu, X.-P. Enhancement of Toyocamycin Production Through Increasing Supply of Precursor GTP in Streptomyces diastatochromogenes 1628. Basic Microbiol. 2022, 62, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Wu, W.; Zhang, F.-X.; Chen, L.; Wang, R.-D.; Ye, J.; Wu, H.-Z.; Zhang, H.-Z. AdpAlin Regulates Lincomycin and Melanin Biosynthesis by Modulating Precursors Flux in Streptomyces lincolnensis. Basic Microbiol. 2023, 63, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Lombó, F.; Pfeifer, B.; Leaf, T.; Ou, S.; Kim, Y.-S.; Cane, D.-E.; Licari, P.; Khosla, C. Enhancing the Atom Economy of Polyketide Biosynthetic Processes through Metabolic Engineering. Biotechnol. Prog. 2001, 17, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-H.; Shen, W.-H.; Yuan, J.-F.; Sun, J.-R.; Wang, M.-Y. Advances in the Biosynthesis of Natamycin and its Regulatory Mechanisms. Chin. J. Biotechnol. 2021, 37, 1107–1119. [Google Scholar]
- Wang, W.-S.; Li, S.-S.; Li, Z.-L.; Zhang, J.-Y.; Fan, K.-Q.; Tan, G.-Y.; Ai, G.-M.; Lam, S.-M.; Shui, G.-H.; Yang, Z.-H.; et al. Harnessing the Intracellular Triacylglycerols for Titer Improvement of Polyketides in Streptomyces. Nat. Biotechnol. 2020, 38, 76–83. [Google Scholar] [CrossRef]
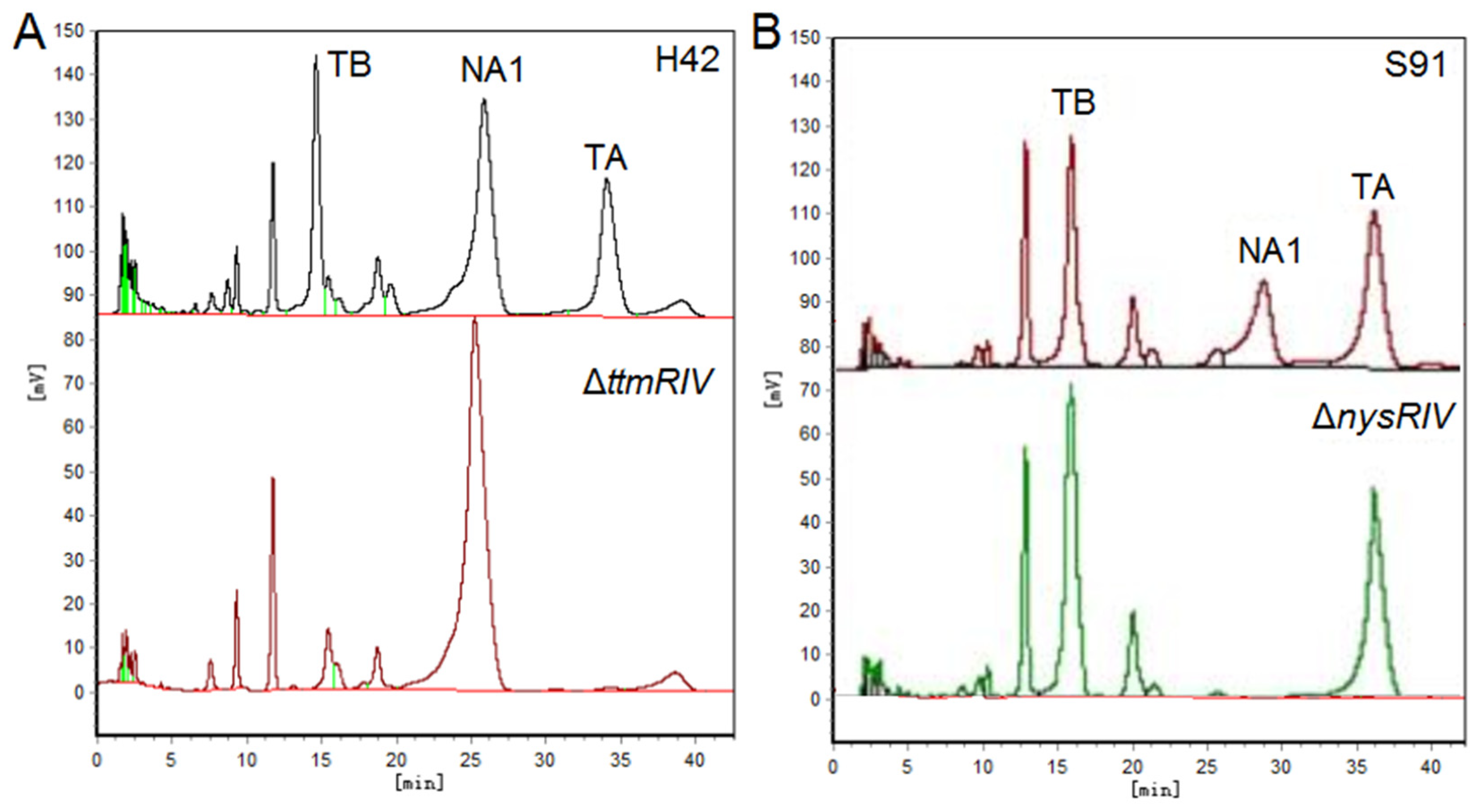
- Ren, J.; Cui, Y.-Q.; Zhang, F.; Cui, H.; Ni, X.-P.; Chen, F.; Li, L.; Xia, H.-Z. Enhancement of Nystatin Production by Redirecting Precursor Fluxes After Disruption of the Tetramycin Gene from Streptomyces ahygroscopicus. Microbiol. Res. 2014, 169, 602–608. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, Y.-Y.; Ran, X.-X.; Fan, W.-M.; Jiang, X.-H.; Guan, W.-J.; Li, Y.-Q. Improvement of Natamycin Production by Engineering of Phosphopantetheinyl Transferases in Streptomyces chattanoogensis L10. Appl. Environ. Microbiol. 2013, 79, 3346–3354. [Google Scholar] [CrossRef]
- Kim, S.-H.; Lee, H.-N.; Kim, H.-J.; Kim, E.-S. Transcriptome Analysis of an Antibiotic Downregulator Mutant and Synergistic Actinorhodin Stimulation Via Disruption of a Precursor Flux Regulator in Streptomyces coelicolor. Appl. Environ. Microbiol. 2011, 77, 1872–1877. [Google Scholar] [CrossRef]
- Li, S.-S.; Li, Z.-L.; Pang, S.; Xiang, W.-S.; Wang, W.-S. Coordinating Precursor Supply for Pharmaceutical Polyketide Production in Streptomyces. Curr. Opin. Biotechnol. 2021, 69, 26–34. [Google Scholar] [CrossRef]
- Ahn, S.-K.; Cuthbertson, L.; Nodwell, J.-R. Genome Context As a Predictive Tool for Identifying Regulatory Targets of the TetR Family Transcriptional Regulators. PLoS ONE 2012, 7, e50562. [Google Scholar] [CrossRef] [PubMed]
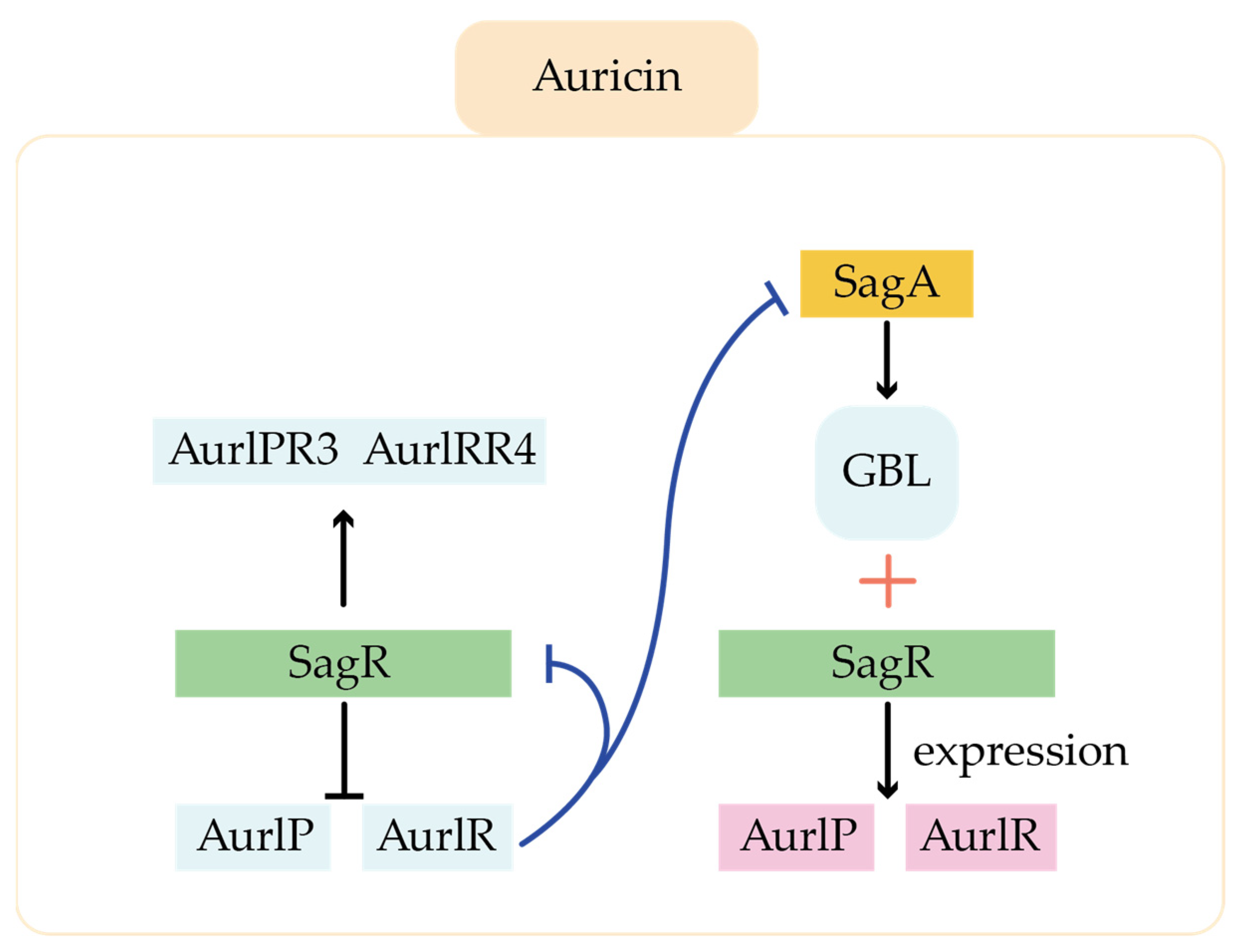
- Mingyar, E.; Feckova, L.; Novakova, R.; Bekova, C.; Kormanec, J. A γ-butyrolactone Autoregulator-Receptor System Involved in the Regulation of Auricin Production in Streptomyces aureofaciens CCM 3239. Appl. Microbiol. Biotechnol. 2014, 99, 309–325. [Google Scholar] [CrossRef]
- Du, Y.-L.; Shen, X.-L.; Yu, P.; Bai, L.-Q.; Li, Y.-Q. Gamma-Butyrolactone Regulatory System of Streptomyces chattanoogensis Links Nutrient Utilization, Metabolism, and Development. Appl. Environ. Microbiol. 2011, 77, 8415–8426. [Google Scholar] [CrossRef]
- Chen, J.-S.; Liu, M.; Liu, X.-T.; Miao, J.; Fu, C.-Z.; Gao, H.-Y.; Müller, R.; Zhang, Q.; Zhang, L.-X. Interrogation of Streptomyces avermitilis for Efficient Production of Avermectins. Synth. Syst. Biotechnol. 2016, 1, 7–16. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Yan, T.-T.; Jiang, L.-B.; Wen, Y.; Song, Y.; Chen, Z.; Li, J.-L. Characterization of SAV7471, ATetR-Family Transcriptional Regulator Involved in the Regulation of Coenzyme A Metabolism in Streptomyces avermitilis. J. Bacteriol. 2013, 195, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Liu, W.-S.; Sun, D.; Luo, S.; Chen, Z.; Wen, Y.; Li, J.-L. Engineering of the TetR Family Transcriptional Regulator SAV151 and its Target Genes Increases Avermectin Production in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 2013, 98, 399–409. [Google Scholar] [CrossRef] [PubMed]
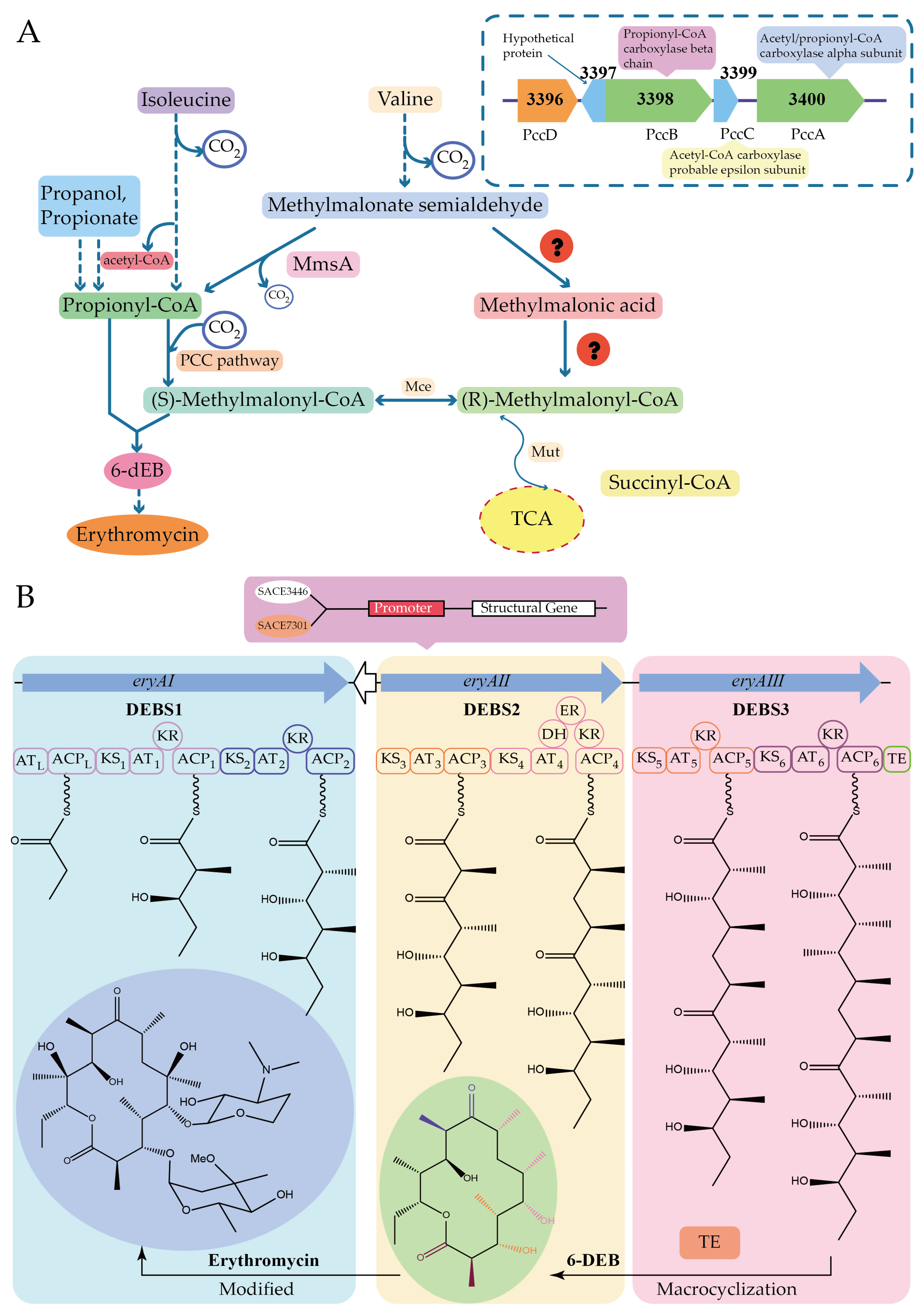
- Xu, Z.; Wang, M.-M.; Ye, B.-C. TetR Family Transcriptional Regulator PccD Negatively Controls Propionyl Coenzyme A Assimilation in Saccharopolyspora erythraea. J. Bacteriol. 2017, 199, 00281-17. [Google Scholar] [CrossRef]
- Wu, H.; Chen, M.; Mao, Y.-R.; Li, W.-W.; Liu, J.-T.; Huang, X.-D.; Zhou, Y.; Ye, B.-C.; Zhang, L.-X.; Weaver, D.; et al. Dissecting and Engineering of the TetR Family Regulator SACE_7301 For Enhanced Erythromycin Production in Saccharopolyspora erythraea. Microb. Cell Fact. 2014, 13, 158. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Y.-S.; Yuan, L.; Mao, Y.-R.; Wang, W.-W.; Zhu, L.; Wu, P.-P.; Fu, C.-Z.; Müller, R.; Weaver, D.-T.; et al. Inactivation of SACE_3446, A TetR Family Transcriptional Regulator, Stimulates Erythromycin Production in Saccharopolyspora erythraea. Synth. Syst. Biotechnol. 2016, 1, 39–46. [Google Scholar] [CrossRef]
- Irzik, K.; Van Ooyen, J.; Gätgens, J.; Krumbach, K.; Bott, M.; Eggeling, L. Acyl-CoA Sensing by FasR to Adjust Fatty Acid Synthesis in Corynebacterium glutamicum. J. Biotechnol. 2014, 192, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.-F.; Ramos, A.; Liras, P. Regulation of Geldanamycin Biosynthesis by Cluster-Situated Transcription Factors and the Master Regulator PhoP. Antibiotics 2019, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.-X.; Yin, M.; Wu, S.-H.; Han, X.-L.; Ji, K.-Y.; Wen, M.-L.; Lu, T. GdmRIII, A TetR Family Transcriptional Regulator, Controls Geldanamycin and Elaiophylin Biosynthesis in Streptomyces autolyticus CGMCC0516. Sci. Rep. 2017, 7, 4803. [Google Scholar] [CrossRef] [PubMed]
- Lyu, M.; Cheng, Y.-Q.; Han, X.; Wen, Y.; Song, Y.; Li, J.-L.; Chen, Z. AccR, a TetR Family Transcriptional Repressor, Coordinates Short-Chain Acyl Coenzyme A Homeostasis in Streptomyces avermitilis. Appl. Environ. Microbiol. 2020, 86, e00508-20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-W.; Feng, R.-R.; Zheng, G.-S.; Tian, J.-Z.; Ruan, L.-J.; Ge, M.; Jiang, W.-H.; Lu, Y.-H. Involvement of the TetR-Type Regulator PaaR in the Regulation of Pristinamycin I Biosynthesis through an Effect on Precursor Supply in Streptomyces pristinaespiralis. J. Bacteriol. 2015, 197, 2062–2071. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.-L.; Jia, X.; Xiao, Y. Accumulation of Malonyl-CoA in E.coli Promoted by CRISPRi/ddCpf1. J. Food Sci. Biotechnol. 2021, 40, 17–25. [Google Scholar]
- Ji, X.-Y.; Zhao, H.-W.; Zhu, H.; Zhu, K.; Tang, S.-Y.; Lou, C.-B. CRISPRi/dCpf1-mediated Dynamic Metabolic Switch to Enhance Butenoic Acid Production in Escherichia coli. Appl. Microbiol. Biotechnol. 2020, 104, 5385–5393. [Google Scholar] [CrossRef]
- Li, M.-Y.; Chen, J.-Z.; Wang, Y.; Liu, J.; Huang, J.-W.; Chen, N.; Zheng, P.; Sun, J.-B. Efficient Multiplex Gene Repression by CRISPR-dCpf1 in Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2020, 24, 357. [Google Scholar] [CrossRef]
- Li, J.; Mu, X.; Dong, W.-Y.; Chen, Y.; Kang, Q.-J.; Zhao, G.; Hou, J.; Gonzalez, R.; Bai, L.-Q.; Feng, Y.; et al. A Non-carboxylative Route for the Efficient Synthesis of Central Metabolite Malonyl-CoA and its Derived Products. Nat. Catal. 2024, 1–14. [Google Scholar] [CrossRef]


| Chemical Compound | Precursor |
|---|---|
| short-chain fatty acid | acetic acid, propionic acid, malonic acid, methylmalonic acid, butyric acid |
| isoprene | isopentenyl pyrophosphate |
| amino acids and aromatic intermediates | protein amino acid |
| sugar and amino sugar | butylose, hexose, pentose, aminosugar |
| cyclohexanol and aminocyclohexanol | actinomycin, inositol, inosine, streptoguanidine |
| amidinopyridine | argnine |
| purine and pyrimidine bases | adenine, guanine, cytosine |
| aromatic intermediates and aromatic amino acids | mangiferolic acid, branching acid, tyrosine, tryptophan |
| methyl group | S-adenosylmethionine |
| Precursor | Product | Regulator | Microorganisms | Binding Site Sequences a | Reference |
|---|---|---|---|---|---|
| acyl-CoA | avermectin | SAV7471 | S.avermitilis | 5′-GAGAACSWWCGTTCTC-3′ | [39] |
| – | avermectin | SAV151 | S.avermitilis | 5′-GAACTGACACTCTAGCTTGTCAGTTC-3′ 5′-TCTGACATGGTGTC GTGTCAGA-3′ | [40] |
| propionyl-CoA | erythromycin | PccD | Saccharopolyspora erythraea | 5′-T/ATGACGG/CTGT/CTGT/A-3′ | [41] |
| acetyl-CoA carboxylase | fatty acids | FasR | Corynebacterium glutamic | 5′-AAAANATGACNANNTCCTCATNTTT-3′ | [44] |
| malonyl-CoA methoxylmalonyl-CoA | geldanamycin elaiophylin | GdmRIII | S. autolyticus | 5′-CACCATGATGGAGGACCACT-3′ 5′-AGGCCATCGAGGACTGGCTG-3′ | [45,46] |
| isobutyryl-CoA | avermectin | AccR | S.avermitilis A14 | 5′-GTTAA–N6–TTAAC-3′ | [47] |
| L-phenylglycine | pristinamycin I | PaaR | S. pristinaespiralis | 5′-AACGA-N4-TCGGT-3′ b | [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, X.; Dong, Y.; Gu, Y.; Cui, H. Effect of Precursors and Their Regulators on the Biosynthesis of Antibiotics in Actinomycetes. Molecules 2024, 29, 1132. https://doi.org/10.3390/molecules29051132
Yan X, Dong Y, Gu Y, Cui H. Effect of Precursors and Their Regulators on the Biosynthesis of Antibiotics in Actinomycetes. Molecules. 2024; 29(5):1132. https://doi.org/10.3390/molecules29051132
Chicago/Turabian StyleYan, Xu, Yao Dong, Yawen Gu, and Hao Cui. 2024. "Effect of Precursors and Their Regulators on the Biosynthesis of Antibiotics in Actinomycetes" Molecules 29, no. 5: 1132. https://doi.org/10.3390/molecules29051132
APA StyleYan, X., Dong, Y., Gu, Y., & Cui, H. (2024). Effect of Precursors and Their Regulators on the Biosynthesis of Antibiotics in Actinomycetes. Molecules, 29(5), 1132. https://doi.org/10.3390/molecules29051132