Unraveling the Dynamics of Host–Microbiota Indole Metabolism: An Investigation of Indole, Indolin-2-one, Isatin, and 3-Hydroxyindolin-2-one
Abstract
:1. Introduction
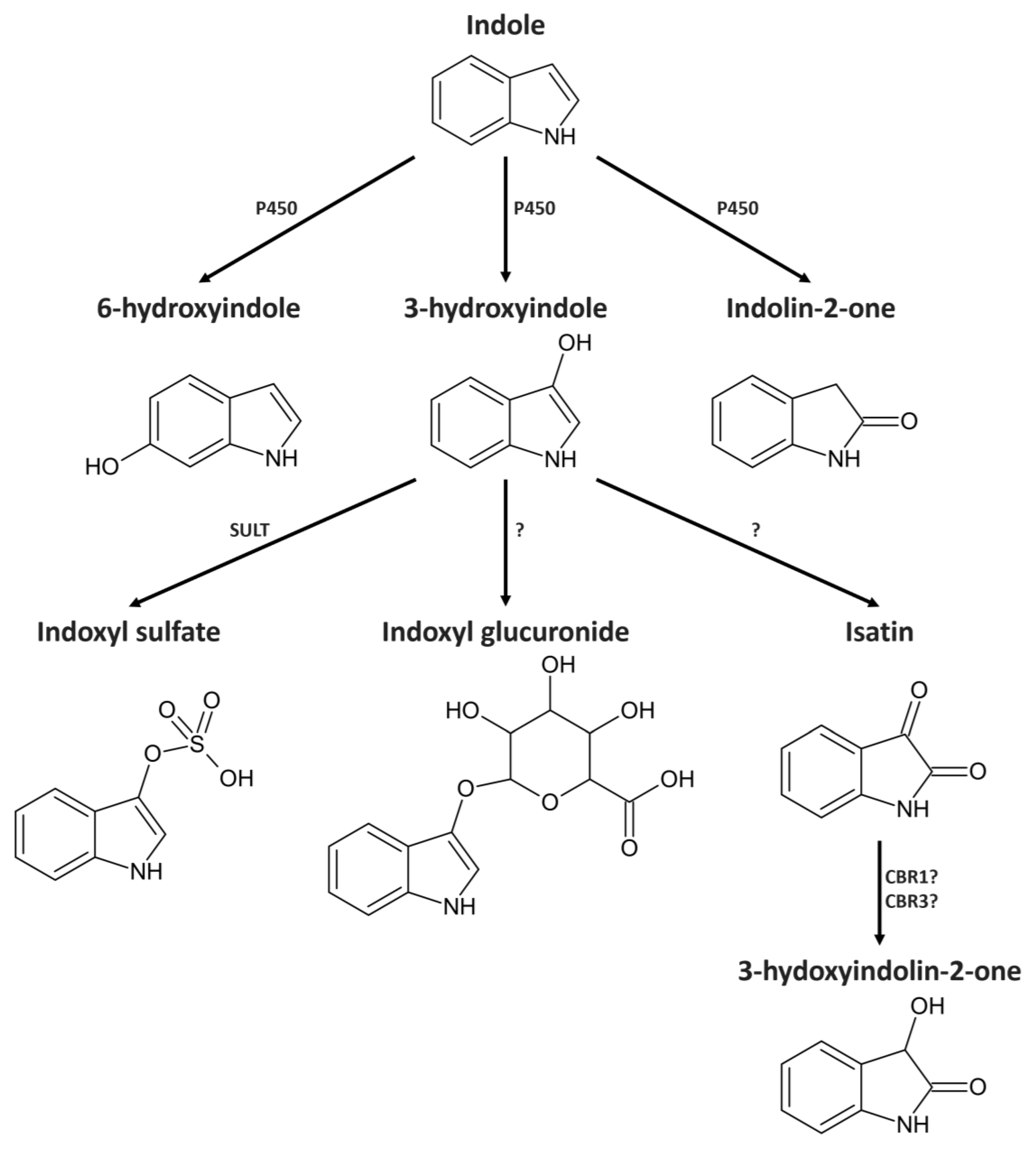
2. Results
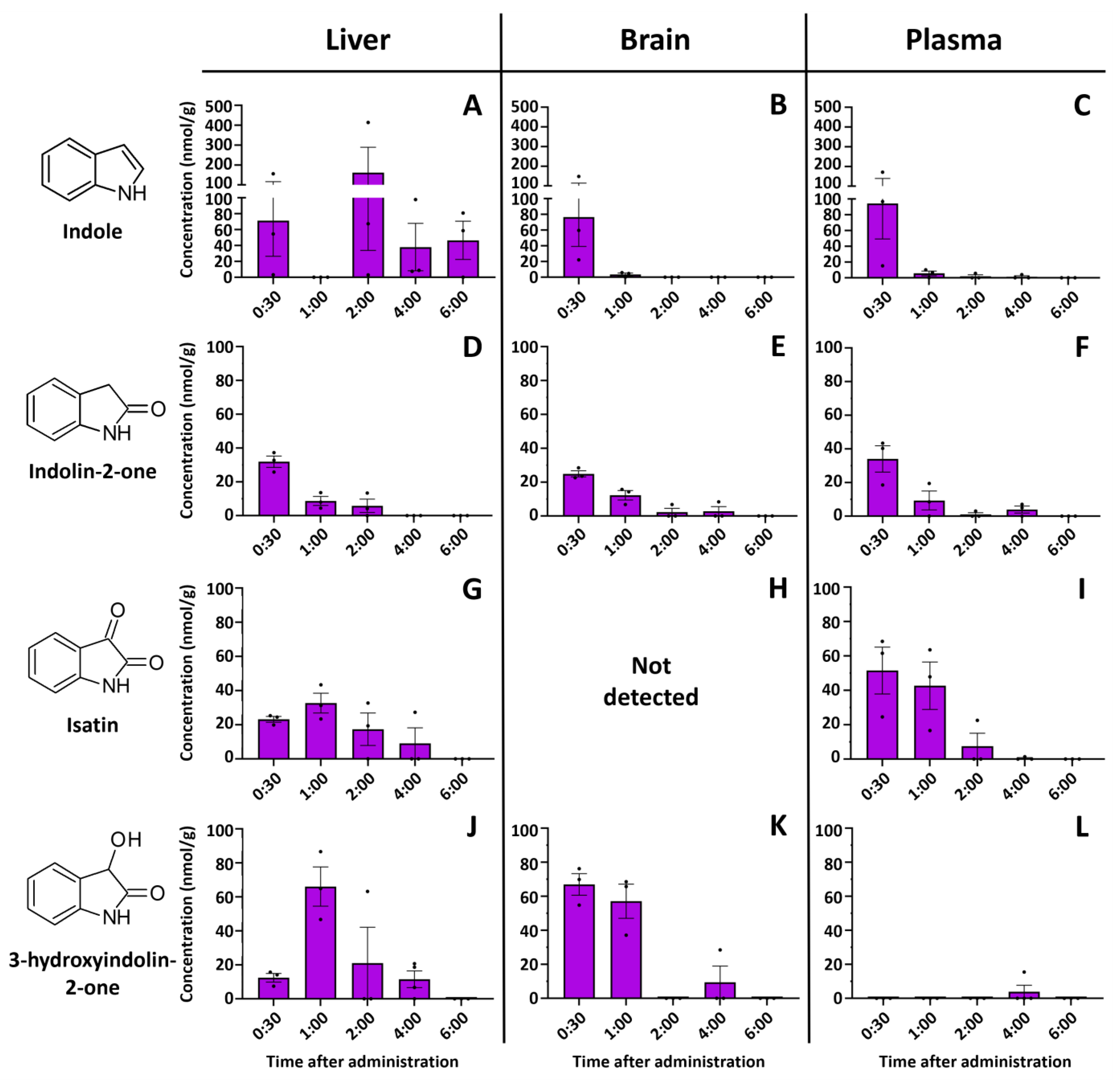
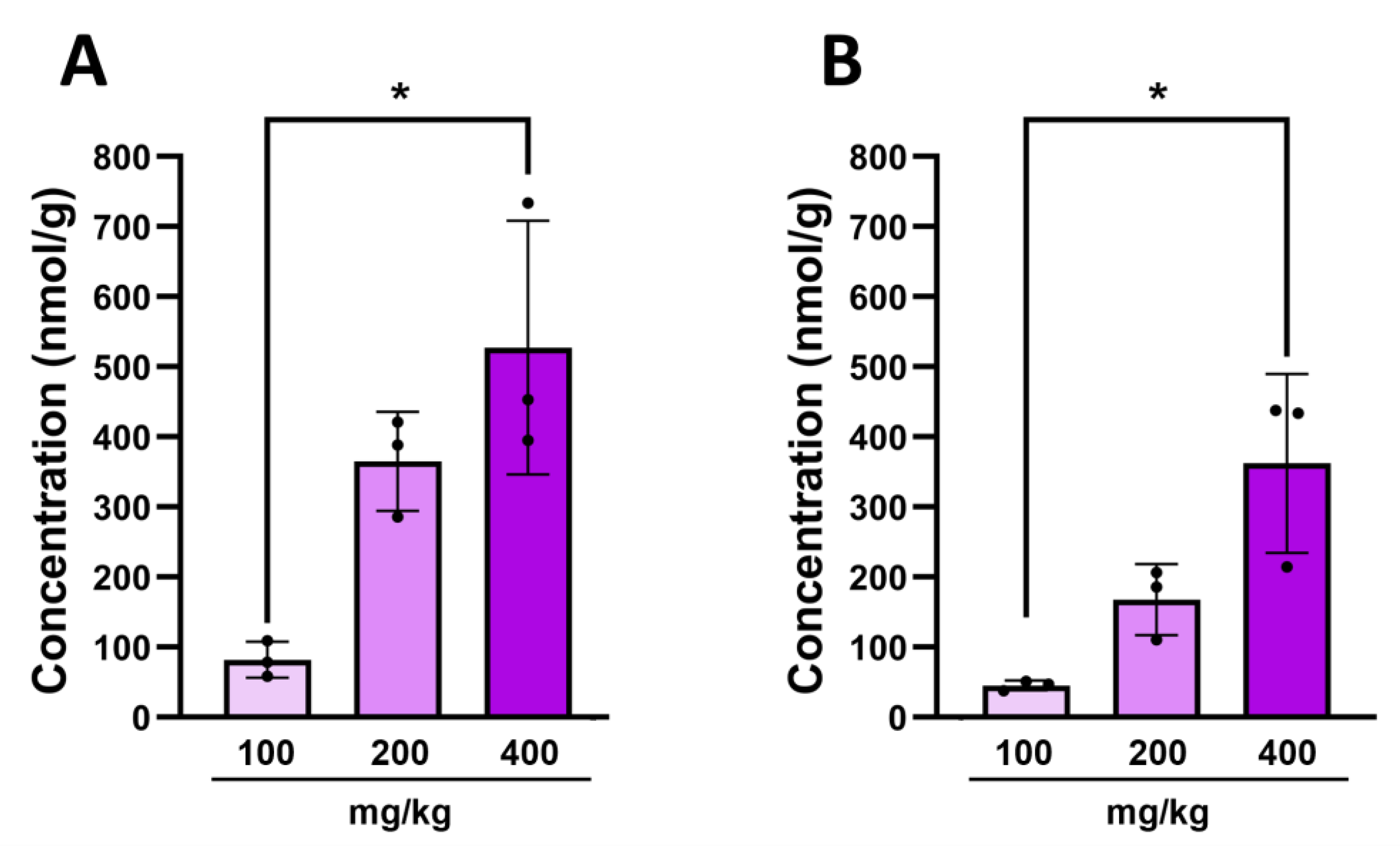
2.1. Indole Is Quickly and Efficiently Metabolized In Vivo
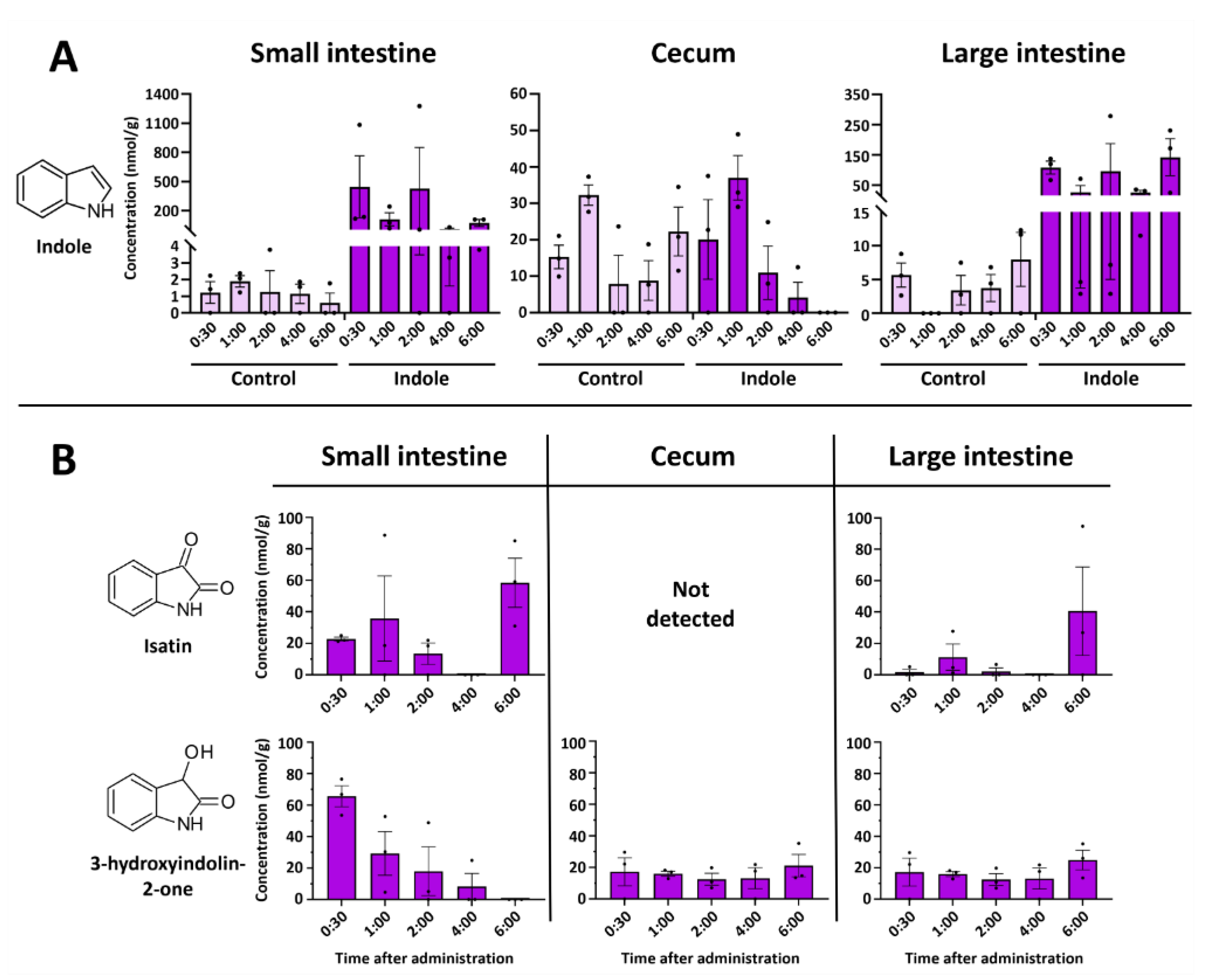
2.2. Indole from the Digestive Tract Is Metabolized by the Host and Produces Multiple Bioactive Metabolites
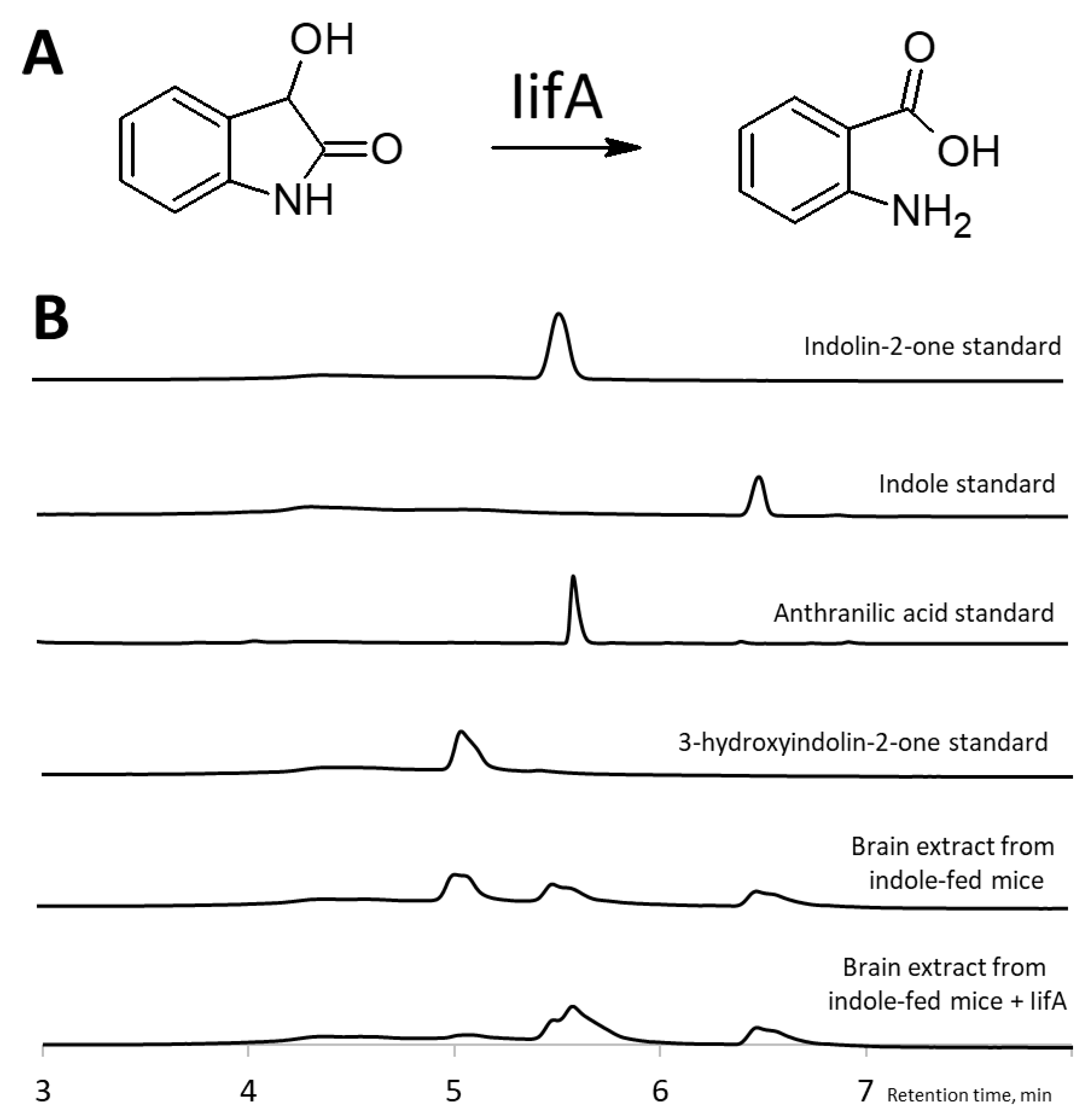
2.3. Identification of 3-Hydroxyindolin-2-one as Indole Metabolite
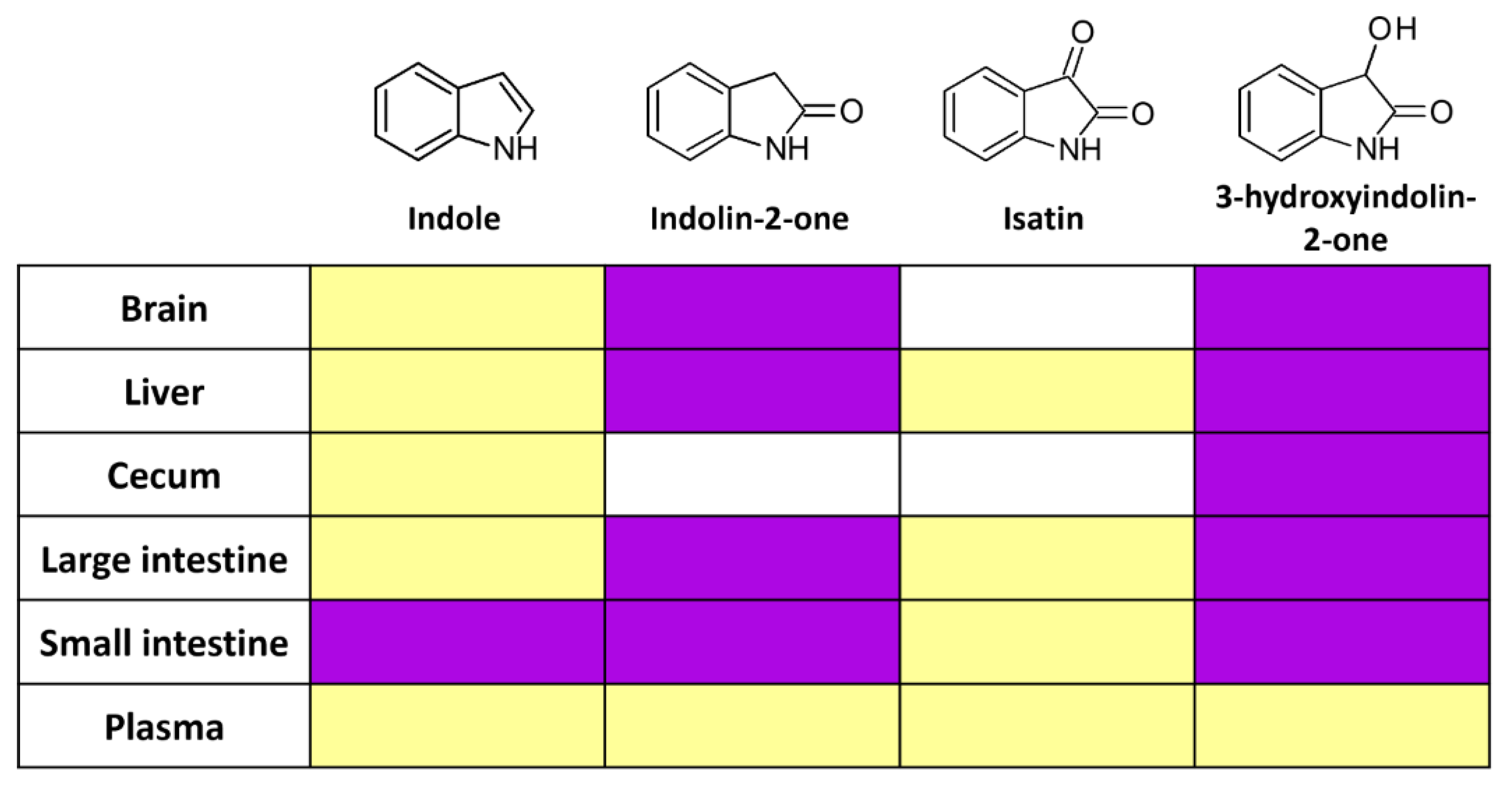
2.4. Intestinal 3-Hydroxyindolin-2-one Can Cross the Blood–Brain Barrier
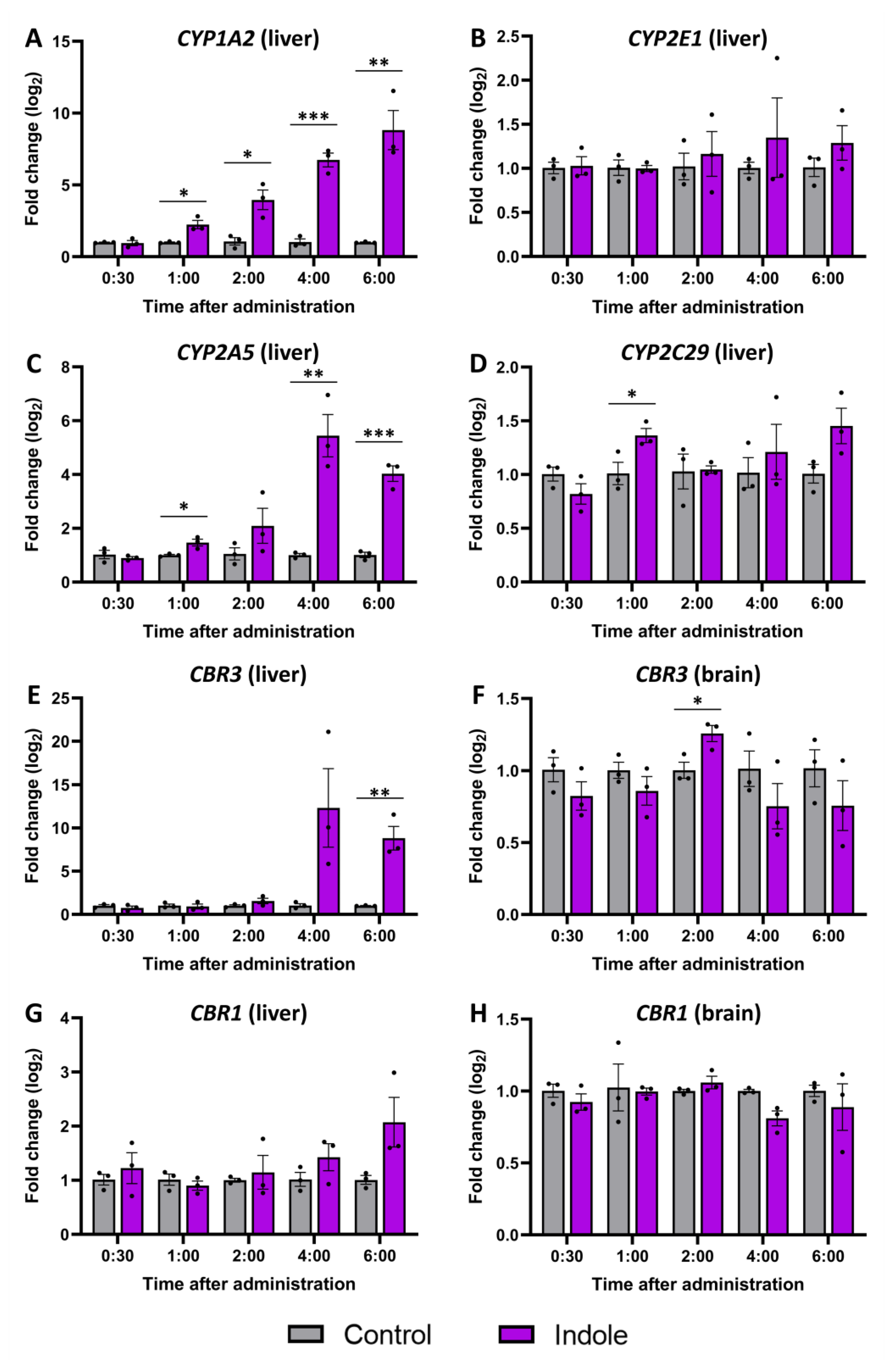
2.5. Gene Expression Changes Caused by Indole Administration
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animals
4.3. Administration of Indole and 3-Hydroxyindolin-2-one
4.4. Liquid–Liquid Extraction of Indole and Its Metabolites
4.5. Quantification of Indole Metabolites by HPLC/MS
4.6. Gene Expression Analysis by Real-Time PCR (RT-PGR)
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, B.; Wang, X.; Li, L. Human Gut Microbiome: The Second Genome of Human Body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Megur, A.; Baltriukiene, D.; Bukelskiene, V.; Burokas, A. The Microbiota–Gut–Brain Axis and Alzheimer’s Disease: Neuroinflammation Is to Blame? Nutrients 2020, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Burokas, A.; Moloney, R.; Dinan, T.; Cryan, J. Microbiota Regulation of the Mammalian Gut–Brain Axis. Adv. Appl. Microbiol. 2015, 91, 1–62. [Google Scholar]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the Normal Gut Microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the Gut Microbiota in Nutrition and Health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef]
- Cai, H.; Chen, X.; Burokas, A.; Maldonado, R. Editorial: Gut Microbiota as a Therapeutic Target in Neuropsychiatric Disorders: Current Status and Future Directions. Front. Neurosci. 2023, 17, 1198291. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lee, J. Indole as an Intercellular Signal in Microbial Communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef] [PubMed]
- Golubeva, A.V.; Joyce, S.A.; Moloney, G.; Burokas, A.; Sherwin, E.; Arboleya, S.; Flynn, I.; Khochanskiy, D.; Moya-Pérez, A.; Peterson, V.; et al. Microbiota-Related Changes in Bile Acid & Tryptophan Metabolism Are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 2017, 24, 166–178. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial Tryptophan Catabolites in Health and Disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Hyland, N.P.; Cavanaugh, C.R.; Hornby, P.J. Emerging Effects of Tryptophan Pathway Metabolites and Intestinal Microbiota on Metabolism and Intestinal Function. Amino Acids 2022, 54, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Kamath, A.V.; Vaidyanathan, C.S. New Pathway for the Biodegradation of Indole in Aspergillus Niger. Appl. Environ. Microbiol. 1990, 56, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhang, C.; Mu, Y.; Shen, Q.; Feng, Y. Indole Affects Biofilm Formation in Bacteria. Indian J. Microbiol. 2010, 50, 362–368. [Google Scholar] [CrossRef]
- Hirakawa, H.; Hayashi-Nishino, M.; Yamaguchi, A.; Nishino, K. Indole Enhances Acid Resistance in Escherichia coli. Microb. Pathog. 2010, 49, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Russell, R.M.; Hoskan, M.A.; Sperandio, V. Indole Sensing Regulator (IsrR) Promotes Virulence Gene Expression in Enteric Pathogens. mBio 2022, 13, e01939-22. [Google Scholar] [CrossRef] [PubMed]
- Field, C.M.; Summers, D.K. Indole Inhibition of ColE1 Replication Contributes to Stable Plasmid Maintenance. Plasmid 2012, 67, 88–94. [Google Scholar] [CrossRef]
- Hendrikx, T.; Schnabl, B. Indoles: Metabolites Produced by Intestinal Bacteria Capable of Controlling Liver Disease Manifestation. J. Intern. Med. 2019, 286, 32–40. [Google Scholar] [CrossRef]
- Kim, J.; Park, W. Indole: A Signaling Molecule or a Mere Metabolic Byproduct That Alters Bacterial Physiology at a High Concentration? J. Microbiol. 2015, 53, 421–428. [Google Scholar] [CrossRef]
- Ye, X.; Li, H.; Anjum, K.; Zhong, X.; Miao, S.; Zheng, G.; Liu, W.; Li, L. Dual Role of Indoles Derived From Intestinal Microbiota on Human Health. Front. Immunol. 2022, 13, 903526. [Google Scholar] [CrossRef]
- Sadauskas, M.; Vaitekūnas, J.; Gasparavičiūtė, R.; Meškys, R. Indole Biodegradation in Acinetobacter sp. Strain O153: Genetic and Biochemical Characterization. Appl. Environ. Microbiol. 2017, 83, e01453-17. [Google Scholar] [CrossRef]
- Tennoune, N.; Andriamihaja, M.; Blachier, F. Production of Indole and Indole-Related Compounds by the Intestinal Microbiota and Consequences for the Host: The Good, the Bad, and the Ugly. Microorganisms 2022, 10, 930. [Google Scholar] [CrossRef] [PubMed]
- Gillam, E.M.J.; Notley, L.M.; Cai, H.; De Voss, J.J.; Guengerich, F.P. Oxidation of Indole by Cytochrome P450 Enzymes. Biochemistry 2000, 39, 13817–13824. [Google Scholar] [CrossRef] [PubMed]
- Banoglu, E.; Jha, G.G.; King, R.S. Hepatic Microsomal Metabolism of Indole to Indoxyl, a Precursor of Indoxyl Sulfate. Eur. J. Drug Metab. Pharmacokinet. 2001, 26, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Jaglin, M.; Rhimi, M.; Philippe, C.; Pons, N.; Bruneau, A.; Goustard, B.; Daugé, V.; Maguin, E.; Naudon, L.; Rabot, S. Indole, a Signaling Molecule Produced by the Gut Microbiota, Negatively Impacts Emotional Behaviors in Rats. Front. Neurosci. 2018, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Carpenedo, R.; Mannaioni, G.; Moroni, F. Oxindole, a Sedative Tryptophan Metabolite, Accumulates in Blood and Brain of Rats with Acute Hepatic Failure. J. Neurochem. 1998, 70, 1998–2003. [Google Scholar] [CrossRef] [PubMed]
- Glover, V.; Bhattacharya, S.K.; Chakrabarti, A.; Sandler, M. The Psychopharmacology of Isatin: A Brief Review. Stress Med. 1998, 14, 225–229. [Google Scholar] [CrossRef]
- Cho, H.-U.; Kim, S.; Sim, J.; Yang, S.; An, H.; Nam, M.-H.; Jang, D.-P.; Lee, C.J. Redefining Differential Roles of MAO-A in Dopamine Degradation and MAO-B in Tonic GABA Synthesis. Exp. Mol. Med. 2021, 53, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, A.; Buneeva, O.; Glover, V. Biological Targets for Isatin and Its Analogues: Implications for Therapy. Biologics 2007, 1, 151–162. [Google Scholar]
- Satou, R.; Izumikawa, M.; Katsuyama, Y.; Matsui, M.; Takagi, M.; Shin-ya, K.; Ohnishi, Y. Isolation, Structural Elucidation and Biosynthesis of 3-Hydroxy-6-Dimethylallylindolin-2-One, a Novel Prenylated Indole Derivative from Actinoplanes Missouriensis. J. Antibiot. 2014, 67, 231–236. [Google Scholar] [CrossRef]
- Spiteller, P.; Glawischnig, E.; Gierl, A.; Steglich, W. Studies on the Biosynthesis of 2-Hydroxy-1,4-Benzoxazin-3-One (HBOA) from 3-Hydroxyindolin-2-One in Zea Mays. Phytochemistry 2001, 57, 373–376. [Google Scholar] [CrossRef]
- Leong, S.C.; Sirich, T.L. Indoxyl Sulfate—Review of Toxicity and Therapeutic Strategies. Toxins 2016, 8, 358. [Google Scholar] [CrossRef]
- Usami, N.; Kitahara, K.; Ishikura, S.; Nagano, M.; Sakai, S.; Hara, A. Characterization of a Major Form of Human Isatin Reductase and the Reduced Metabolite. Eur. J. Biochem./FEBS 2001, 268, 5755–5763. [Google Scholar] [CrossRef]
- Beloborodova, N.V.; Chernevskaya, E.A.; Getsina, M.L. Indolic Structure Metabolites as Potential Biomarkers of Non-Infectious Diseases. Curr. Pharm. Des. 2021, 27, 238–249. [Google Scholar] [CrossRef]
- Bian, J.; Sun, J.; Chang, H.; Wei, Y.; Cong, H.; Yao, M.; Xiao, F.; Wang, H.; Zhao, Y.; Liu, J.; et al. Profile and Potential Role of Novel Metabolite Biomarkers, Especially Indoleacrylic Acid, in Pathogenesis of Neuromyelitis Optica Spectrum Disorders. Front. Pharmacol. 2023, 14, 1166085. [Google Scholar] [CrossRef]
- Liu, Y.; Pei, Z.; Pan, T.; Wang, H.; Chen, W.; Lu, W. Indole Metabolites and Colorectal Cancer: Gut Microbial Tryptophan Metabolism, Host Gut Microbiome Biomarkers, and Potential Intervention Mechanisms. Microbiol. Res. 2023, 272, 127392. [Google Scholar] [CrossRef]
- Abel, E.L. Behavioral Effects of Isatin on Open Field Activity and Immobility in the Forced Swim Test in Rats. Physiol. Behav. 1995, 57, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, H.; Hu, J.; Zheng, J.; Zhou, J.; Botchlett, R.; Matthews, D.; Zeng, T.; Chen, L.; Xiao, X.; et al. Indole Alleviates Diet-Induced Hepatic Steatosis and Inflammation in a Manner Involving Myeloid Cell 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3. Hepatology 2020, 72, 1191–1203. [Google Scholar] [CrossRef]
- Balcz, B.; Kirchner, L.; Cairns, N.; Fountoulakis, M.; Lubec, G. Increased Brain Protein Levels of Carbonyl Reductase and Alcohol Dehydrogenase in Down Syndrome and Alzheimer’s Disease. In Protein Expression in Down Syndrome Brain; Lubec, G., Ed.; Springer: Vienna, Austria, 2001; pp. 193–201. [Google Scholar] [CrossRef]
- Zhang, X.; Qu, Y.; Ma, Q.; Kong, C.; Zhou, H.; Cao, X.; Shen, W.; Shen, E.; Zhou, J. Production of Indirubin from Tryptophan by Recombinant Escherichia Coli Containing Naphthalene Dioxygenase Genes from Comamonas Sp. MQ. Appl. Biochem. Biotechnol. 2014, 172, 3194–3206. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Wang, K.; Zhang, P.; Ren, S.; Sun, J.; Yang, M.; Zhang, F.; Sun, B. Carbonyl Reductase 1 Attenuates Ischemic Brain Injury by Reducing Oxidative Stress and Neuroinflammation. Transl. Stroke Res. 2021, 12, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.-W.; Kim, S.D.; Rhee, M.H.; Shin, J.-H. Pharmacological Actions of 5-Hydroxyindolin-2 on Modulation of Platelet Functions and Thrombus Formation via Thromboxane A2 Inhibition and cAMP Production. Int. J. Mol. Sci. 2022, 23, 14545. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kunevičius, A.; Sadauskas, M.; Raudytė, J.; Meškys, R.; Burokas, A. Unraveling the Dynamics of Host–Microbiota Indole Metabolism: An Investigation of Indole, Indolin-2-one, Isatin, and 3-Hydroxyindolin-2-one. Molecules 2024, 29, 993. https://doi.org/10.3390/molecules29050993
Kunevičius A, Sadauskas M, Raudytė J, Meškys R, Burokas A. Unraveling the Dynamics of Host–Microbiota Indole Metabolism: An Investigation of Indole, Indolin-2-one, Isatin, and 3-Hydroxyindolin-2-one. Molecules. 2024; 29(5):993. https://doi.org/10.3390/molecules29050993
Chicago/Turabian StyleKunevičius, Arnas, Mikas Sadauskas, Julija Raudytė, Rolandas Meškys, and Aurelijus Burokas. 2024. "Unraveling the Dynamics of Host–Microbiota Indole Metabolism: An Investigation of Indole, Indolin-2-one, Isatin, and 3-Hydroxyindolin-2-one" Molecules 29, no. 5: 993. https://doi.org/10.3390/molecules29050993
APA StyleKunevičius, A., Sadauskas, M., Raudytė, J., Meškys, R., & Burokas, A. (2024). Unraveling the Dynamics of Host–Microbiota Indole Metabolism: An Investigation of Indole, Indolin-2-one, Isatin, and 3-Hydroxyindolin-2-one. Molecules, 29(5), 993. https://doi.org/10.3390/molecules29050993





