Characterization of an Aptamer Targeting Neu5Gc, as an Endogenous Pathogenic Factor Derived from Red Meat
Abstract
:1. Introduction
2. Results
2.1. Production and Analysis of the Solid Phase Antigen Neu5Gc-BSA
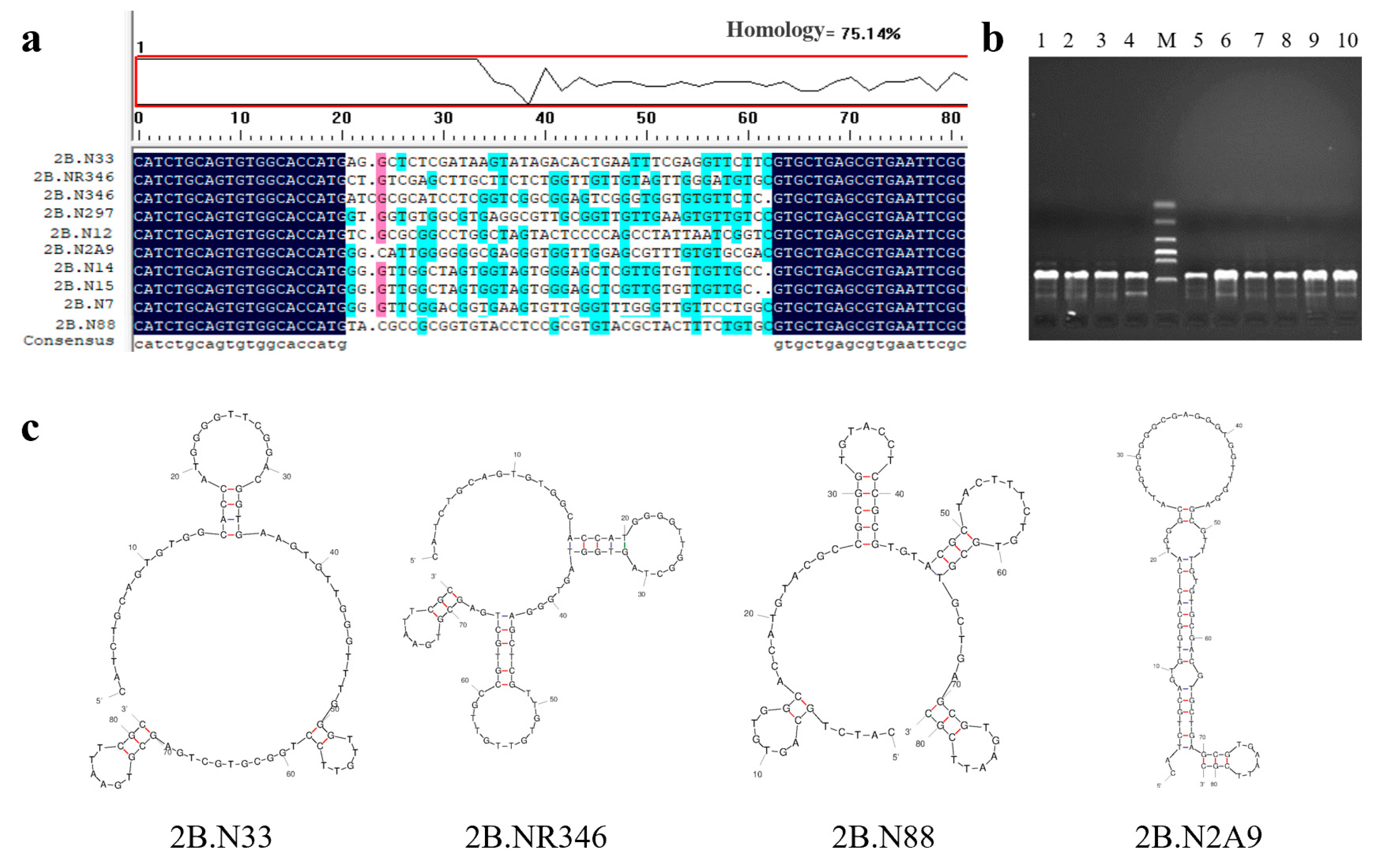
2.2. Sequence Analysis of Nucleic Acid Aptamers
2.3. Affinity Measurement Results
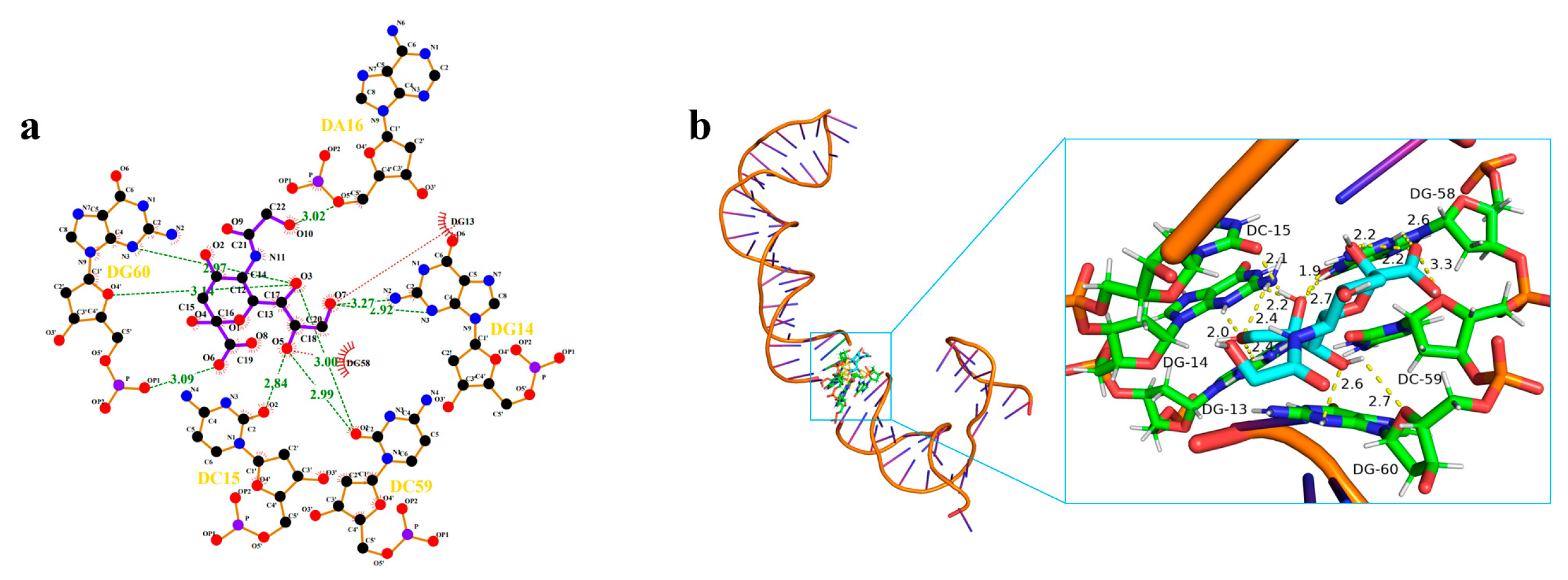
2.4. Analysis of the Binding Mechanism
2.5. pH Stability and Specificity Measurements
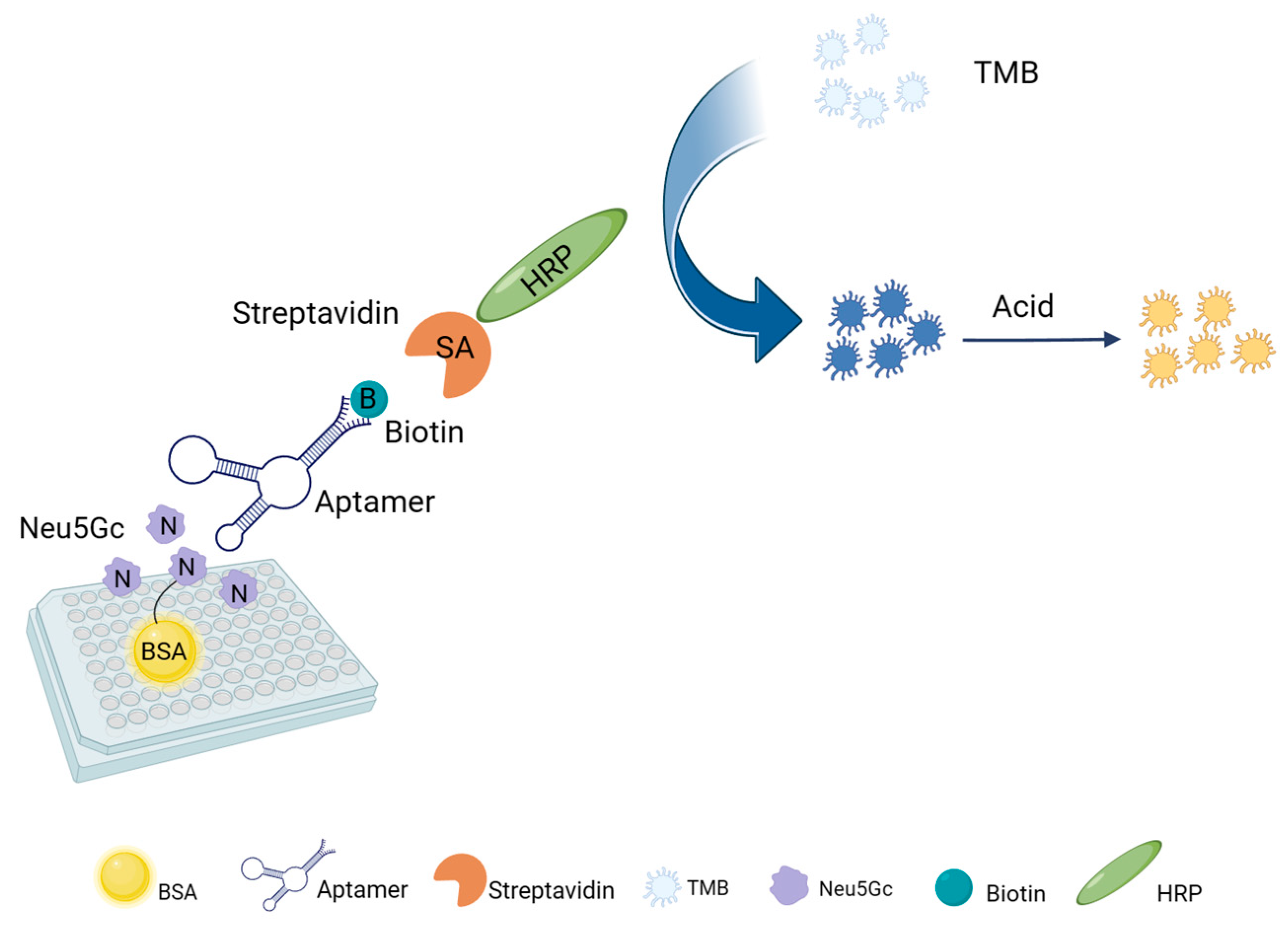
2.6. Aptamer-Based ELOSA Assay Feasibility Analysis
2.7. Feasibility Analysis of Aptamers in the Detection of Red Meat Samples
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Neu5Gc-BSA Conjugate
4.3. The Acquisition of Nucleic Acid Aptamers and Analysis of Predictions
4.4. Determination of Binding Affinity for Candidate Aptamers
4.5. Molecular Docking Methods
4.6. The Evaluation of pH Stability and Specificity of Aptamers
4.7. Establishment of an ELOSA Assay for Neu5Gc Based on Selected Aptamer
4.8. Preparation and Analysis of Red Meat Samples
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Varki, A. Advances in the Biology and Chemistry of Sialic Acids. ACS Chem. Biol. 2010, 5, 163–176. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. (Eds.) Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Wang, B. Sialic Acid Is an Essential Nutrient for Brain Development and Cognition. Annu. Rev. Nutr. 2009, 29, 177–222. [Google Scholar] [CrossRef]
- Kim, A.E.; Lundgreen, A.; Wolff, R.K.; Fejerman, L.; John, E.M.; Torres-Mejía, G.; Ingles, S.A.; Boone, S.D.; Connor, A.E.; Hines, L.M.; et al. Red Meat, Poultry, and Fish Intake and Breast Cancer Risk among Hispanic and Non-Hispanic White Women: The Breast Cancer Health Disparities Study. Cancer Causes Control 2016, 27, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Je, Y. Meat Consumption and Risk of Metabolic Syndrome: Results from the Korean Population and a Meta-Analysis of Observational Studies. Nutrients 2018, 10, 390. [Google Scholar] [CrossRef]
- Zhao, Z.; Yin, Z.; Zhao, Q. Red and Processed Meat Consumption and Gastric Cancer Risk: A Systematic Review and Meta-Analysis. Oncotarget 2017, 8, 30563–30575. [Google Scholar] [CrossRef] [PubMed]
- Pearce, O.M.T.; Läubli, H.; Verhagen, A.; Secrest, P.; Zhang, J.; Varki, N.M.; Crocker, P.R.; Bui, J.D.; Varki, A. Inverse Hormesis of Cancer Growth Mediated by Narrow Ranges of Tumor-Directed Antibodies. Proc. Natl. Acad. Sci. USA 2014, 111, 5998–6003. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xie, B.; Wang, B.; Troy, F.A. LC-MS/MS Glycomic Analyses of Free and Conjugated Forms of the Sialic Acids, Neu5Ac, Neu5Gc and KDN in Human Throat Cancers. Glycobiology 2015, 25, 1362–1374. [Google Scholar] [CrossRef]
- Liu, J.; Cao, Z.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef]
- Das, R.; Dhiman, A.; Kapil, A.; Bansal, V.; Sharma, T.K. Aptamer-Mediated Colorimetric and Electrochemical Detection of Pseudomonas Aeruginosa Utilizing Peroxidase-Mimic Activity of Gold NanoZyme. Anal. Bioanal. Chem. 2019, 411, 1229–1238. [Google Scholar] [CrossRef]
- Tao, W.; Zeng, X.; Wu, J.; Zhu, X.; Yu, X.; Zhang, X.; Zhang, J.; Liu, G.; Mei, L. Polydopamine-Based Surface Modification of Novel Nanoparticle-Aptamer Bioconjugates for In Vivo Breast Cancer Targeting and Enhanced Therapeutic Effects. Theranostics 2016, 6, 470–484. [Google Scholar] [CrossRef]
- Wang, S.; Yong, W.; Liu, J.; Zhang, L.; Chen, Q.; Dong, Y. Development of an Indirect Competitive Assay-Based Aptasensor for Highly Sensitive Detection of Tetracycline Residue in Honey. Biosens. Bioelectron. 2014, 57, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Abnous, K.; Danesh, N.M.; Sarreshtehdar Emrani, A.; Ramezani, M.; Taghdisi, S.M. A Novel Fluorescent Aptasensor Based on Silica Nanoparticles, PicoGreen and Exonuclease III as a Signal Amplification Method for Ultrasensitive Detection of Myoglobin. Anal. Chim. Acta 2016, 917, 71–78. [Google Scholar] [CrossRef]
- Gong, S.; Ren, H.-L.; Tian, R.-Y.; Lin, C.; Hu, P.; Li, Y.-S.; Liu, Z.-S.; Song, J.; Tang, F.; Zhou, Y.; et al. A Novel Analytical Probe Binding to a Potential Carcinogenic Factor of N-Glycolylneuraminic Acid by SELEX. Biosens. Bioelectron. 2013, 49, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Ren, H.; Lin, C.; Hu, P.; Tian, R.; Liu, Z.; Li, Y.; Zhou, Y.; Yang, Y.; Lu, S. Immunochromatographic Strip Biosensor for the Rapid Detection of N-Glycolylneuraminic Acid Based on Aptamer-Conjugated Nanoparticle. Anal. Biochem. 2018, 561–562, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Z.; Li, H.; Liu, J.; Xie, G.; Xiao, F.; Wu, X.; Aguilar, Z.P.; Xu, H. A Competitive Enzyme Linked Aptasensor with Rolling Circle Amplification (ELARCA) Assay for Colorimetric Detection of Listeria Monocytogenes. Food Control 2020, 107, 106806. [Google Scholar] [CrossRef]
- Wang, S.; Dong, Y.; Liang, X. Development of a SPR Aptasensor Containing Oriented Aptamer for Direct Capture and Detection of Tetracycline in Multiple Honey Samples. Biosens. Bioelectron. 2018, 109, 1–7. [Google Scholar] [CrossRef]
- Xu, X.; Ying, Y.; Li, Y. One-Step and Label-Free Detection of Alpha-Fetoprotein Based on Aggregation of Gold Nanorods. Sens. Actuators B Chem. 2012, 175, 194–200. [Google Scholar] [CrossRef]
- AOAC INTERNATIONAL. Available online: https://www.aoac.org/ (accessed on 6 March 2024).
- Yao, H.L.; Conway, L.P.; Wang, M.M.; Huang, K.; Liu, L.; Voglmeir, J. Quantification of Sialic Acids in Red Meat by UPLC-FLD Using Indoxylsialosides as Internal Standards. Glycoconj. J. 2016, 33, 219–226. [Google Scholar] [CrossRef]
- Yehuda, S.; Padler-Karavani, V. Glycosylated Biotherapeutics: Immunological Effects of N-Glycolylneuraminic Acid. Front. Immunol. 2020, 11, 21. [Google Scholar] [CrossRef]
- Grant, W.B. Long Follow-Up Times Weaken Observational Diet–Cancer Study Outcomes: Evidence from Studies of Meat and Cancer Risk. Nutrients 2024, 16, 26. [Google Scholar] [CrossRef]
- Obukhova, P.; Tsygankova, S.; Chinarev, A.; Shilova, N.; Nokel, A.; Kosma, P.; Bovin, N. Are There Specific Antibodies against Neu5Gc Epitopes in the Blood of Healthy Individuals? Glycobiology 2020, 30, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Soulillou, J.-P.; Cozzi, E.; Bach, J.-M. Challenging the Role of Diet-Induced Anti-Neu5Gc Antibodies in Human Pathologies. Front. Immunol. 2020, 11, 834. [Google Scholar] [CrossRef]
- Bashir, S.; Fezeu, L.K.; Leviatan Ben-Arye, S.; Yehuda, S.; Reuven, E.M.; Szabo de Edelenyi, F.; Fellah-Hebia, I.; Le Tourneau, T.; Imbert-Marcille, B.M.; Drouet, E.B.; et al. Association between Neu5Gc Carbohydrate and Serum Antibodies against It Provides the Molecular Link to Cancer: French NutriNet-Santé Study. BMC Med. 2020, 18, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Yu, S.; Wang, W.; Sun, R.; Wu, Z.; Gao, Z.; Pang, Y.; Li, Q. Lamprey Immunity Protein Enables Detection for Bladder Cancer through Recognizing N-Hydroxyacetylneuraminic Acid (Neu5Gc)-Modified as a Diagnostic Marker and Exploration of Its Production Mechanism. Biochem. Biophys. Res. Commun. 2022, 614, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shewell, L.K.; Day, C.J.; Jennings, M.P. N-Glycolylneuraminic Acid as a Carbohydrate Cancer Biomarker. Transl. Oncol. 2023, 31, 101643. [Google Scholar] [CrossRef]
- Lin, X.; Yao, H.; Guo, J.; Huang, Y.; Wang, W.; Yin, B.; Li, X.; Wang, T.; Li, C.; Xu, X.; et al. Protein Glycosylation and Gut Microbiota Utilization Can Limit the In Vitro and In Vivo Metabolic Cellular Incorporation of Neu5Gc. Mol. Nutr. Food Res. 2022, 66, 2100615. [Google Scholar] [CrossRef]
- Manz, C.; Mancera-Arteu, M.; Zappe, A.; Hanozin, E.; Polewski, L.; Giménez, E.; Sanz-Nebot, V.; Pagel, K. Determination of Sialic Acid Isomers from Released N-Glycans Using Ion Mobility Spectrometry. Anal. Chem. 2022, 94, 13323–13331. [Google Scholar] [CrossRef]
- Mastrangeli, R.; Audino, M.C.; Palinsky, W.; Broly, H.; Bierau, H. Current Views on N-Glycolylneuraminic Acid in Therapeutic Recombinant Proteins. Trends Pharmacol. Sci. 2021, 42, 943–956. [Google Scholar] [CrossRef]
- Zhu, Y.; Hart, G.W. Dual-Specificity RNA Aptamers Enable Manipulation of Target-Specific O-GlcNAcylation and Unveil Functions of O-GlcNAc on β-Catenin. Cell 2023, 186, 428–445.e27. [Google Scholar] [CrossRef]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. 2021, 60, 16800–16823. [Google Scholar] [CrossRef]
- Chen, L.; Yang, G.; Qu, F. Advances of Aptamer-Based Small-Molecules Sensors in Body Fluids Detection. Talanta 2024, 268, 125348. [Google Scholar] [CrossRef]
- Bashir, S.; Leviatan Ben Arye, S.; Reuven, E.M.; Yu, H.; Costa, C.; Galiñanes, M.; Bottio, T.; Chen, X.; Padler-Karavani, V. Presentation Mode of Glycans Affect Recognition of Human Serum Anti-Neu5Gc IgG Antibodies. Bioconjugate Chem. 2019, 30, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Hurum, D.C.; Rohrer, J.S. Five-Minute Glycoprotein Sialic Acid Determination by High-Performance Anion Exchange Chromatography with Pulsed Amperometric Detection. Anal. Biochem. 2011, 419, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; van der Molen, J.; Kuipers, F.; van Leeuwen, S.S. Quantitation of Bioactive Components in Infant Formulas: Milk Oligosaccharides, Sialic Acids and Corticosteroids. Food Res. Int. 2023, 174, 113589. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhu, B.; Chen, Z.; Jia, L.; Sun, S. Precision Characterization of Site-Specific O-Acetylated Sialic Acids on N-Glycoproteins. Anal. Chem. 2023, 95, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- Lacomba, R.; Salcedo, J.; Alegría, A.; Jesús Lagarda, M.; Barberá, R.; Matencio, E. Determination of Sialic Acid and Gangliosides in Biological Samples and Dairy Products: A Review. J. Pharm. Biomed. Anal. 2010, 51, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Vos, G.M.; Weber, J.; Sweet, I.R.; Hooijschuur, K.C.; Sastre Toraño, J.; Boons, G.-J. Oxidative Release of O-Glycans under Neutral Conditions for Analysis of Glycoconjugates Having Base-Sensitive Substituents. Anal. Chem. 2023, 95, 8825–8833. [Google Scholar] [CrossRef]
- Safferthal, M.; Bechtella, L.; Zappe, A.; Vos, G.M.; Pagel, K. Labeling of Mucin-Type O-Glycans for Quantification Using Liquid Chromatography and Fluorescence Detection. ACS Meas. Sci. Au 2024. [Google Scholar] [CrossRef]
- Vos, G.M.; Hooijschuur, K.C.; Li, Z.; Fjeldsted, J.; Klein, C.; de Vries, R.P.; Toraño, J.S.; Boons, G.-J. Sialic Acid O-Acetylation Patterns and Glycosidic Linkage Type Determination by Ion Mobility-Mass Spectrometry. Nat. Commun. 2023, 14, 1–13. [Google Scholar] [CrossRef]
- Jahan, M.; Thomson, P.C.; Wynn, P.C.; Wang, B. Red Meat Derived Glycan, N-Acetylneuraminic Acid (Neu5Ac) Is a Major Sialic Acid in Different Skeletal Muscles and Organs of Nine Animal Species—A Guideline for Human Consumers. Foods 2023, 12, 337. [Google Scholar] [CrossRef]
- Jahan, M.; Thomson, P.C.; Wynn, P.C.; Wang, B. The Non-Human Glycan, N-Glycolylneuraminic Acid (Neu5Gc), Is Not Expressed in All Organs and Skeletal Muscles of Nine Animal Species. Food Chem. 2021, 343, 128439. [Google Scholar] [CrossRef]
- Shewell, L.K.; Day, C.J.; Hippolite, T.; De Bisscop, X.; Paton, J.C.; Paton, A.W.; Jennings, M.P. Serum Neu5Gc Biomarkers Are Elevated in Primary Cutaneous Melanoma. Biochem. Biophys. Res. Commun. 2023, 642, 162–166. [Google Scholar] [CrossRef]
- Shewell, L.K.; Day, C.J.; Kutasovic, J.R.; Abrahams, J.L.; Wang, J.; Poole, J.; Niland, C.; Ferguson, K.; Saunus, J.M.; Lakhani, S.R.; et al. N-Glycolylneuraminic Acid Serum Biomarker Levels Are Elevated in Breast Cancer Patients at All Stages of Disease. BMC Cancer 2022, 22, 334. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, C.S.; Kang, M.; Moon, C.; Kim, M.; Yang, S.; Jang, L.; Jang, J.Y.; Jeong, C.M.; Lee, H.S.; et al. Structural Identification and Quantification of Unreported Sialylated N-Glycans in Bovine Testicular Hyaluronidase by LC-ESI-HCD-MS/MS. J. Pharm. Biomed. Anal. 2024, 240, 115938. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Yang, D.-P.; Ye, D.; Zhang, X.; Fang, X.; Zhang, S.; Liu, B.; Kong, J. Protein-Inorganic Hybrid Nanoflowers as Ultrasensitive Electrochemical Cytosensing Interfaces for Evaluation of Cell Surface Sialic Acid. Biosens. Bioelectron. 2015, 68, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Han, E.; Ding, L.; Ju, H. Highly Sensitive Fluorescent Analysis of Dynamic Glycan Expression on Living Cells Using Glyconanoparticles and Functionalized Quantum Dots. Anal. Chem. 2011, 83, 7006–7012. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Chen, J.; Chen, X.; Wang, X.; Zhang, Y.; Zhou, N. Systematic Screening and Optimization of Single-Stranded DNA Aptamer Specific for N-Acetylneuraminic Acid: A Comparative Study. Sens. Actuators B Chem. 2021, 344, 130270. [Google Scholar] [CrossRef]
- Ma, P.; Ye, H.; Guo, H.; Ma, X.; Yue, L.; Wang, Z. Aptamer Truncation Strategy Assisted by Molecular Docking and Sensitive Detection of T-2 Toxin Using SYBR Green I as a Signal Amplifier. Food Chem. 2022, 381, 132171. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Guo, H.; Duan, N.; Ma, X.; Yue, L.; Gu, Q.; Wang, Z. Label Free Structure-Switching Fluorescence Polarization Detection of Chloramphenicol with Truncated Aptamer. Talanta 2021, 230, 122349. [Google Scholar] [CrossRef]
- Varki, A. Sialic Acids in Human Health and Disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, L.; Liu, N.; Troy, F.A.; Wang, B. LC-MS/MS Quantification of N-Acetylneuraminic Acid, N-Glycolylneuraminic Acid and Ketodeoxynonulosonic Acid Levels in the Urine and Potential Relationship with Dietary Sialic Acid Intake and Disease in 3- to 5-Year-Old Children. Br. J. Nutr. 2014, 111, 332–341. [Google Scholar] [CrossRef]
- Jiang, Y.; Fu, H.; Feng, C.; Wu, J. Effects of Different Treatments on N-glycolyIneuraminic Acid Dissociation in Red Meat. Meat Res. 2015, 29, 52–57. [Google Scholar] [CrossRef]
- Duan, N.; Li, C.; Zhu, X.; Qi, S.; Wang, Z.; Wu, S. Simultaneous Coupled with Separate SELEX for Heterocyclic Biogenic Amine-Specific Aptamers Screening and Their Application in Establishment of an Effective Aptasensor. Sens. Actuators B Chem. 2022, 352, 130985. [Google Scholar] [CrossRef]
- Zha, Y.; Lu, S.; Hu, P.; Ren, H.; Liu, Z.; Gao, W.; Zhao, C.; Li, Y.; Zhou, Y. Dual-Modal Immunosensor with Functionalized Gold Nanoparticles for Ultrasensitive Detection of Chloroacetamide Herbicides. ACS Appl. Mater. Interfaces 2021, 13, 6091–6098. [Google Scholar] [CrossRef]
- Samraj, A.N.; Pearce, O.M.T.; Läubli, H.; Crittenden, A.N.; Bergfeld, A.K.; Banda, K.; Gregg, C.J.; Bingman, A.E.; Secrest, P.; Diaz, S.L.; et al. A Red Meat-Derived Glycan Promotes Inflammation and Cancer Progression. Proc. Natl. Acad. Sci. USA 2015, 112, 542–547. [Google Scholar] [CrossRef]




| Neu5Gc and Its Analogues | Structural Formula | IC50 (ng/mL) | Cross-Reaction Rate (%) |
|---|---|---|---|
| Neu5Gc | 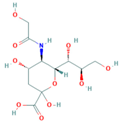 | 9.35 | 100.0% |
| Neu5Ac | 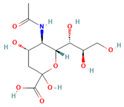 | 467.74 | 2.0% |
| KDN | 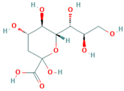 | 1659.58 | 0.56% |
| N-acetyl-D-mannosamine | 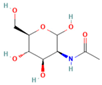 | 36,307.8 | 0.026% |
| Maltose |  | >93,500.00 | <0.01% |
| Sucrose |  | >93,500.00 | <0.01% |
| Glucose |  | >93,500.00 | <0.01% |
| Sample | Free of Neu5Gc (µg/g, Mean ± SD) (n = 3) | Conjugated of Neu5Gc (µg/g, Mean ± SD) (n = 3) | Total of Neu5Gc (µg/g, Mean ± SD) (n = 3) |
|---|---|---|---|
| Beef–Steak | 2.22 ± 0.07 | 21.38 ± 1.04 | 23.60 ± 1.21 |
| Pork–Griskin | 1.217 ± 0.05 | 16.45 ± 1.01 | 17.67 ± 1.03 |
| Mutton–Tenderloin | 1.379 ± 0.22 | 13.95 ± 0.46 | 15.33 ± 0.66 |
| Method | Methodological Features | Reference |
|---|---|---|
| Immunohistochemistry | Polyclonal anti-Neu5Gc chicken IgY was used as an immunogen to recognize gangliosides with the Neu5Gc α2-R terminus. It was applied to immunohistochemistry and TLC overlay. | [34] |
| HPAE-PAD | The sialic acid content in glycolic acid hydrolysate was determined using high-performance anion exchange chromatography with pulsed amperometric detection (HPAE-PAD). Additionally, the separation of Neu5Ac and Neu5Gc in glycolic acid hydrolysate was achieved on an anion exchange column. | [35,36] |
| Mass spectrometry | The signal-to-noise ratio of RP-HPLC tandem MS interfaced with an electrospray ionization source (ESI) for the purification of released Neu5Gc in mass spectrometry and selective ion monitoring can be improved by using its isotopes as internal standards and avoiding derivatization. | [37] |
| Fluorescent marker | The free Neu5Gc was detected through fluorescence labeling with 1,2-diamino-4,5-methylenedioxybenzene (DMB) and subsequent RP-HPLC analysis of the derivative. | [38] |
| Mass Spectrometry (MS) | To facilitate purification and enhance the signal in MAL-DI-TOF mass spectrometry, it is necessary to detect Neu5Gc-carrying glycans by mass spectrometry, permethylate released N- or O-glycan mixtures, and destroy the O-acetyl group through alkaline permethylation for specific release of the glycans. | [39,40,41] |
| Aptamer detection | The Systematic Evolution of Ligands by Exponential Enrichment (SELEX) can be used to select aptamers from chemically synthesized nucleic acid libraries. This method can provide a new analytical probe for developing biosensors to detect Neu5Gc in animal-derived food as well as in the tissues and sera of tumor patients. | [14] |
| Sequence Name | Sequences (5′–3′) | Product Length |
|---|---|---|
| 2B.N33 | CATCTGCAGTGTGGCACCATGGGGTTCGGACGGTGAAGTGTTGGGTTTGGGTTGTTCCTGGCGTGCTGAGCGTGAATTCGC | 81 bp |
| 2B.NR346 | CATCTGCAGTGTGGCACCATGGGGTTGGCTAGTGGTAGTGGGAGCTCGTTGTGTTGTTGCCGTGCTGAGCGTGAATTCGC | 80 bp |
| 2B.N88 | CATCTGCAGTGTGGCACCATGTACGCCGCGGTGTACCTCCGCGTGTACGCTACTTTCTGTGCGTGCTGAGCGTGAATTCGC | 81 bp |
| 2B.N2A9 | CATCTGCAGTGTGGCACCATGGGCATTGGGGGGCGAGGGTGGTTGGAGCGTTTGTGTGCGACGTGCTGAGCGTGAATTCGC | 81 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Ren, H.; Wang, H.; Xiao, Y.; Wang, C.; Liu, M.; Duan, F.; Li, H.; Hu, P.; Li, Y.; et al. Characterization of an Aptamer Targeting Neu5Gc, as an Endogenous Pathogenic Factor Derived from Red Meat. Molecules 2024, 29, 1273. https://doi.org/10.3390/molecules29061273
Guo Y, Ren H, Wang H, Xiao Y, Wang C, Liu M, Duan F, Li H, Hu P, Li Y, et al. Characterization of an Aptamer Targeting Neu5Gc, as an Endogenous Pathogenic Factor Derived from Red Meat. Molecules. 2024; 29(6):1273. https://doi.org/10.3390/molecules29061273
Chicago/Turabian StyleGuo, Yuxi, Honglin Ren, Han Wang, Yiran Xiao, Cong Wang, Mengdi Liu, Fuchun Duan, Haosong Li, Pan Hu, Yansong Li, and et al. 2024. "Characterization of an Aptamer Targeting Neu5Gc, as an Endogenous Pathogenic Factor Derived from Red Meat" Molecules 29, no. 6: 1273. https://doi.org/10.3390/molecules29061273





