The Indiscriminate Chemical Makeup of Secondary Metabolites Derived from Endophytes Harvested from Aloe barbadensis Miller in South Africa’s Limpopo Region
Abstract
:1. Introduction
2. Results
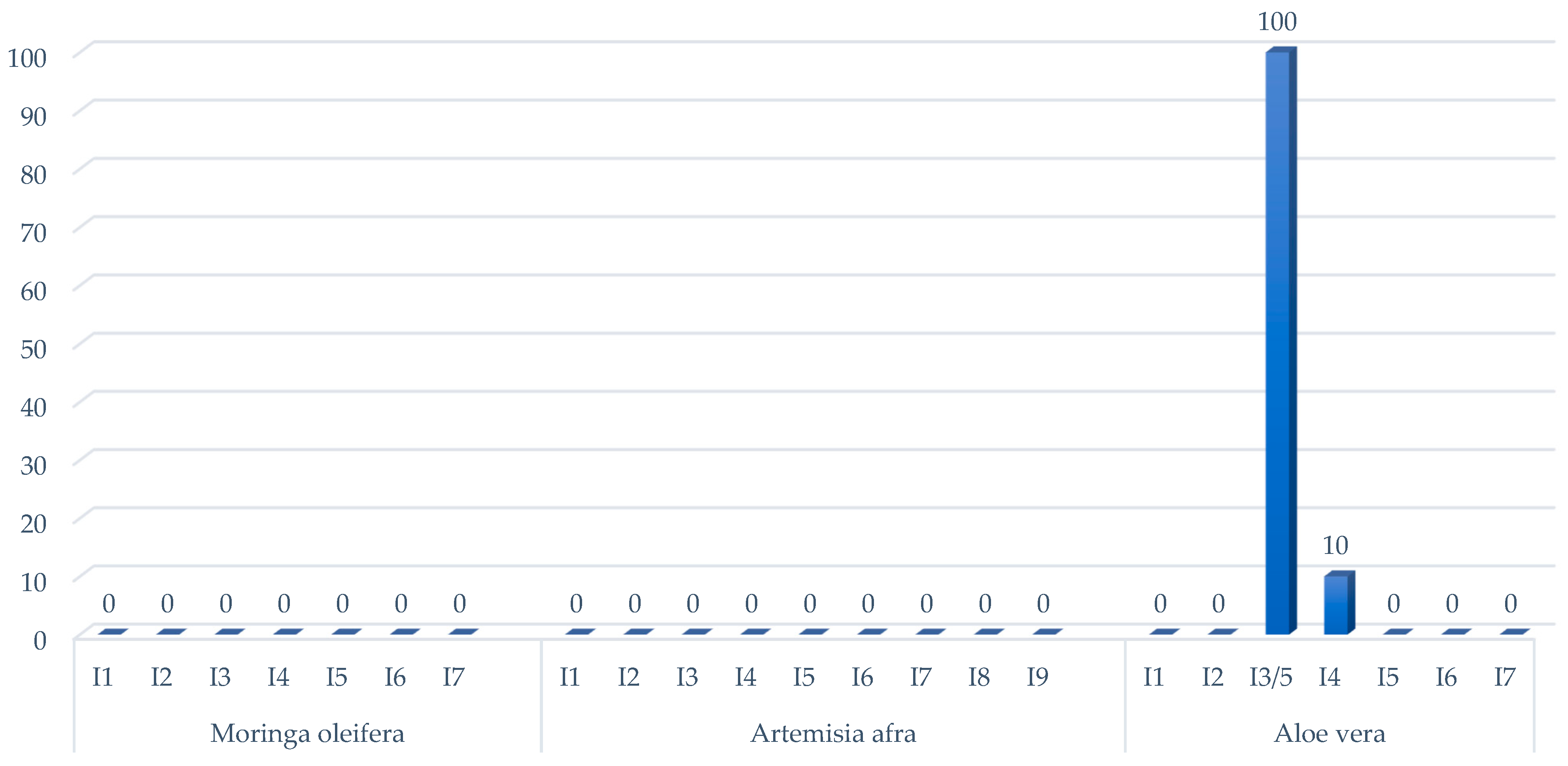
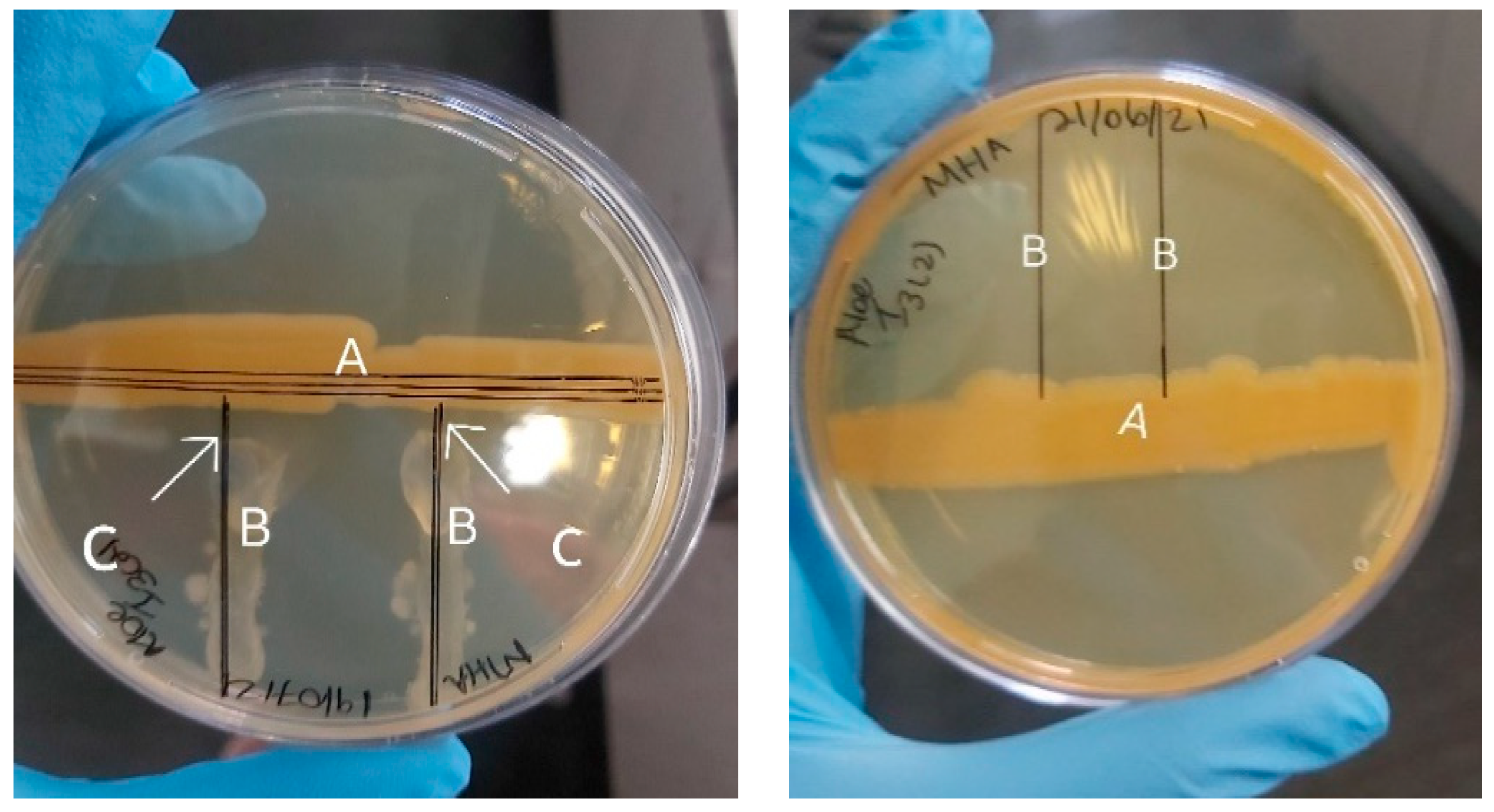
2.1. Antimicrobial Susceptibility of Mycobacterium bovis
2.2. Isolation and Characterization of Endophytes
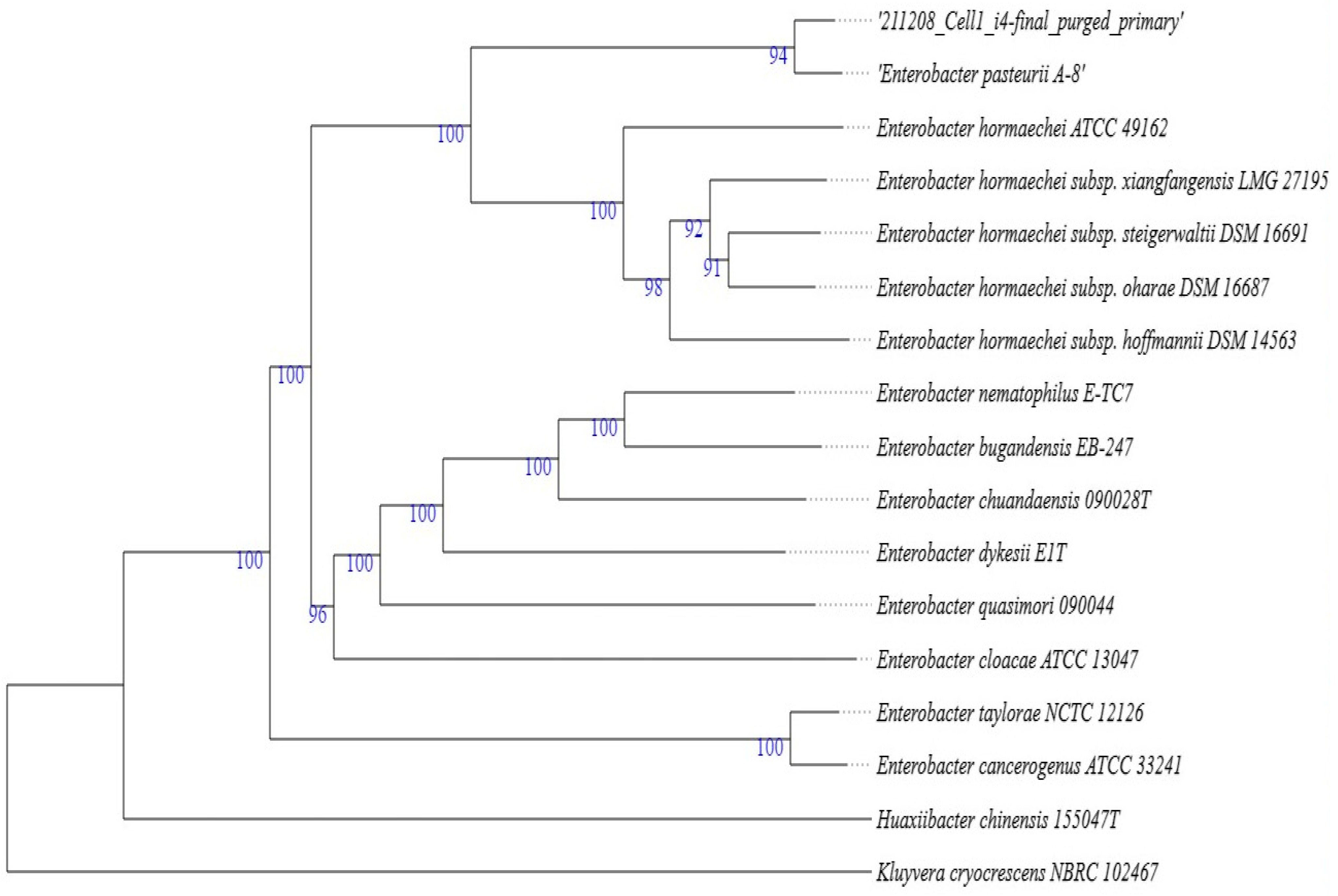
2.3. Molecular and Phylogenetic Identification of Aloe I4 and Aloe I3–I5
2.4. Chemical Profile of Aloe vera I3–I5 and Aloe vera I4
3. Materials and Methods
3.1. Antimicrobial Susceptibility of Mycobacterium bovis against Bacterial Endophytes Isolated from Medicinal Plants
3.1.1. Culturing of Mycobacterium bovis
3.1.2. Susceptibility Test of Mycobacterium bovis against Endophytic Bacteria
3.1.3. Processing of Samples for Metabolite Profiling
3.1.4. Analysis Using GC-HRTOF-MS
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the Sustainable Discovery and Development of New Antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of Microbial Natural Products by Activation of Silent Biosynthetic Gene Clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Agarwal, S.; Verma, K.; Bhardwaj, R.; Mathur, V. Therapeutic Compounds from Medicinal Plant Endophytes: Molecular and Metabolic Adaptations. J. Appl. Microbiol. 2023, 134, lxad074. [Google Scholar] [CrossRef]
- Tsipinana, S.; Husseiny, S.; Alayande, K.A.; Raslan, M.; Amoo, S.; Adeleke, R. Contribution of Endophytes towards Improving Plant Bioactive Metabolites: A Rescue Option against Red-Taping of Medicinal Plants. Front. Plant Sci. 2023, 14, 1248319. [Google Scholar] [CrossRef] [PubMed]
- Cuvas-Limón, R.B.; Ferreira-Santos, P.; Cruz, M.; Teixeira, J.A.; Belmares, R.; Nobre, C. Novel Bio-Functional Aloe vera Beverages Fermented by Probiotic Enterococcus faecium and Lactobacillus lactis. Molecules 2022, 27, 2473. [Google Scholar] [CrossRef]
- Sánchez, M.; González-Burgos, E.; Iglesias, I.; Gómez-Serranillos, M.P. Pharmacological Update Properties of Aloe vera and Its Major Active Constituents. Molecules 2020, 25, 1324. [Google Scholar] [CrossRef]
- Liu Ning, Q.; Cao, M.; Frédérich, M.; Choi, Y.H.; Verpoorte, R.; Van Der Kooy, F. Metabolomic Investigation of the Ethnopharmacological Use of Artemisia afra with NMR Spectroscopy and Multivariate Data Analysis. J. Ethnopharmacol. 2010, 128, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Moyo, P.; Botha, M.E.; Nondaba, S.; Niemand, J.; Maharaj, V.J.; Eloff, J.N.; Louw, A.I.; Birkholtz, L. In Vitro Inhibition of Plasmodium falciparum Early and Late Stage Gametocyte Viability by Extracts from Eight Traditionally Used South African Plant Species. J. Ethnopharmacol. 2016, 185, 235–242. [Google Scholar] [CrossRef]
- Martini, M.C.; Zhang, T.; Williams, J.T.; Abramovitch, R.B.; Weathers, P.J.; Shell, S.S. Artemisia annua and Artemisia afra Extracts Exhibit Strong Bactericidal Activity against Mycobacterium tuberculosis. J. Ethnopharmacol. 2020, 262, 113191. [Google Scholar] [CrossRef]
- Kou, X.; Li, B.; Olayanju, J.; Drake, J.; Chen, N. Nutraceutical or Pharmacological Potential of Moringa oleifera Lam. Nutrients 2018, 10, 343. [Google Scholar] [CrossRef]
- Hashem, A.H.; Al-Askar, A.A.; Abd Elgawad, H.; Abdelaziz, A.M. Bacterial Endophytes from Moringa oleifera Leaves as a Promising Source for Bioactive Compounds. Separations 2023, 10, 395. [Google Scholar] [CrossRef]
- Abdelaziz, A.M.; El-Wakil, D.A.; Hashem, A.H.; Al-Askar, A.A.; AbdElgawad, H.; Attia, M.S. Efficient Role of Endophytic Aspergillus terreus in Biocontrol of Rhizoctonia solani Causing Damping-off Disease of Phaseolus vulgaris and Vicia faba. Microorganisms 2023, 11, 1487. [Google Scholar] [CrossRef]
- Ibáñez, A.; Garrido-Chamorro, S.; Barreiro, C. Microorganisms and Climate Change: A Not so Invisible Effect. Microbiol. Res. 2023, 14, 918–947. [Google Scholar] [CrossRef]
- Damavandi, M.S.; Shojaei, H.; Esfahani, B.N. The Anticancer and Antibacterial Potential of Bioactive Secondary Metabolites Derived from Bacterial Endophytes in Association with Artemisia absinthium. Sci. Rep. 2023, 13, 18473. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Lertcanawanichakul, M.; Sawangnop, S. A Comparison of Two Methods Used for Measuring the Antagonistic Activity of Bacillus Species. Walailak J. Sci. Technol. 2008, 5, 161–171. [Google Scholar]
- Miché, L.; Balandreau, J. Effects of Rice Seed Surface Sterilization with Hypochlorite on Inoculated Burkholderia vietnamiensis. Appl. Environ. Microbiol. 2001, 67, 3046–3052. [Google Scholar] [CrossRef] [PubMed]
- Strobel, G.A. Endophytes as Sources of Bioactive Products. Microbes Infect. 2003, 5, 535–544. [Google Scholar] [CrossRef]
- Xiong, X.Q.; Liao, H.D.; Ma, J.S.; Liu, X.M.; Zhang, L.Y.; Shi, X.W.; Yang, X.L.; Lu, X.N.; Zhu, Y.H. Isolation of a Rice Endophytic Bacterium, Pantoea sp. Sd-1, with Ligninolytic Activity and Characterization of Its Rice Straw Degradation Ability. Lett. Appl. Microbiol. 2014, 58, 123–129. [Google Scholar] [CrossRef]
- Sun, X.; Guo, L.-D. Endophytic Fungal Diversity: Review of Traditional and Molecular Techniques. Mycology 2012, 3, 65–76. [Google Scholar]
- Kim, M.; Chun, J. 16S rRNA Gene-Based Identification of Bacteria and Archaea Using the EzTaxon Server. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 41, pp. 61–74. ISBN 978-0-12-800176-9. [Google Scholar]
- Menpara, D.; Chanda, S. Endophytic Bacteria-Unexplored Reservoir of Antimicrobials for Combating Microbial Pathogens. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Formatex Research Center: Badajoz, Spain, 2013; pp. 1095–1103. [Google Scholar]
- Mezzatesta, M.L.; Gona, F.; Stefani, S. Enterobacter cloacae Complex: Clinical Impact and Emerging Antibiotic Resistance. Future Microbiol. 2012, 7, 887–902. [Google Scholar] [CrossRef]
- Cooney, S.; O’Brien, S.; Iversen, C.; Fanning, S. Bacteria: Other Pathogenic Enterobacteriaceae—Enterobacter and other Genera. In Encyclopedia of Food Safety; Elsevier: Amsterdam, The Netherlands, 2014; pp. 433–441. ISBN 978-0-12-378613-5. [Google Scholar]
- Jasim, B.; Joseph, A.A.; John, C.J.; Mathew, J.; Radhakrishnan, E.K. Isolation and Characterization of Plant Growth Promoting Endophytic Bacteria from the Rhizome of Zingiber officinale. 3 Biotech 2014, 4, 197–204. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural Products: A Continuing Source of Novel Drug Leads. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Hsouna, A.B.; Trigui, M.; Mansour, R.B.; Jarraya, R.M.; Damak, M.; Jaoua, S. Chemical Composition, Cytotoxicity Effect and Antimicrobial Activity of Ceratonia siliqua Essential Oil with Preservative Effects against Listeria Inoculated in Minced Beef Meat. Int. J. Food Microbiol. 2011, 148, 66–72. [Google Scholar] [CrossRef]
- Centres for Disease Control and Prevention. Mycobacterium Bovis (Bovine Tuberculosis) in Humans; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2012. [Google Scholar]
- Larrañaga, M.D.; Lewis, R.J.; Lewis, R.A. Hawley’s Condensed Chemical Dictionary, 16th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; ISBN 978-1-119-31246-8. [Google Scholar]
- Rao, M.R.K.; Anisha, G.; Prabhu, K.; Shil, S.; Vijayalakshmi, N. Preliminary Phytochemical and Gas Chromatography–Mass Spectrometry Study of One Medicinal Plant Carissa carandas. Drug Invent. Today 2019, 12, 1629–1634. [Google Scholar]
- Schmidt, R.; Griesbaum, K.; Behr, A.; Biedenkapp, D.; Voges, H.W.; Garbe, D.; Paetz, C.; Collin, G.; Mayer, D.; Höke, H. Hydrocarbons. Ullmann’s Encyclopedia of Industrial Chemistry, 7th ed.; John Wiley & Sons: New York, NY, USA, 2016. [Google Scholar]
- Vujanovic, S.; Vujanovic, J.; Vujanovic, V. Microbiome-Driven Proline Biogenesis in Plants under Stress: Perspectives for Balanced Diet to Minimize Depression Disorders in Humans. Microorganisms 2022, 10, 2264. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, L.; Li, L.; Zheng, C.; Guo, L.; Li, W.; Sun, P.; Qin, L. Recent Developments and Future Prospects of Antimicrobial Metabolites Produced by Endophytes. Microbiol. Res. 2010, 165, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Okudoh, V.I.; Wallis, F.M. Antimicrobial Activity of Rare Actinomycetes Isolated from Natural Habitats in KwaZulu-Natal, South Africa: Research Articles. S. Afr. J. Sci. 2007, 103, 216–222. [Google Scholar]
- Xu, X.; Legay, S.; Sergeant, K.; Zorzan, S.; Leclercq, C.C.; Charton, S.; Giarola, V.; Liu, X.; Challabathula, D.; Renaut, J.; et al. Molecular Insights into Plant Desiccation Tolerance: Transcriptomics, Proteomics and Targeted Metabolite Profiling in Craterostigma plantagineum. Plant J. 2021, 107, 377–398. [Google Scholar] [CrossRef]
- Daji, G.A.; Green, E.; Akanni, G.B.; Adebo, O.A. GC–MS-based Metabolomics Elucidates the Impact of Cereal-based Microbial Starters on the Metabolite Profile of Mahewu Prepared from Yellow Maize. Int. J. Food Sci. Technol. 2023. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]

| Contig Type | Polished Contigs | Maximum Contig Length | Mean Contig Length | Median Contig Length | N50 Contig Length | Sum of Contig Lengths | e-Size (Sum of Squares | Number of Circular Contigs |
|---|---|---|---|---|---|---|---|---|
| Primary contigs | 3 | 2,598,250 | 1,572,706 | 1,434,460 | 2,598,250 | 4,718,120 | 1,966,539 | 0 |
| Haplotigs | 1 | 10.181 | 10.181 | 10.181 | 10.181 | 10.181 | 10.181 | N/A |
| Contig Type | Polished Contigs | Maximum Contig Length | Mean Contig Length | Median Contig Length | N50 Contig Length | Sum of Contig Lengths | e-Size (Sum of Squares | Number of Circular Contigs |
|---|---|---|---|---|---|---|---|---|
| Primary contigs | 1 | 4,840,834 | 4,840,834 | 4,840,834 | 4,840,834 | 4,840,834 | 4,840,834 | 1 |
| Haplotigs | 3 | 17.724 | 15.141 | 16.977 | 16.977 | 45.425 | 15.792 | N/A |
| R.T. (s) | Area% | Observed Ion m/z | Compound Name | Formula | Area | Similarity | Peak S/N | |
|---|---|---|---|---|---|---|---|---|
| Alcohols | ||||||||
| 1 | 738.27 | 0.035 | 206.166 | 2,4-Di-tert-butylphenol | C14H22O | 117,980 | 888 | 650 |
| 2 | 371.83 | 0.96 | 122.07 | Phenylethyl Alcohol | C8H10O | 3,250,678 | 940 | 3197 |
| 3 | 763.75 | 0.14 | 125.06 | 3-Mercapto-3-methylbutanol | C5H12OS | 479,614 | 785 | 813 |
| 4 | 957.02 | 0.07 | 161.08 | Tryptophol | C10H11NO | 224,164 | 752 | 328 |
| Alkanes and Alkenes | ||||||||
| 5 | 270.83 | 0.14 | 127.65 | Tridecane, 4-methyl- | C14H30 | 454,419.5 | 905.5 | 488.0 |
| 6 | 789.12 | 1.54 | 196.04 | Hexadecane | C16H34 | 5,258,816 | 940 | 3164 |
| 7 | 818.01 | 0.08 | 1671.612 | 2-Dodecanone | C12H24O | 287,327 | 843 | 510 |
| 8 | 832.25 | 0.52 | 161.41 | Pentadecane | C15H32 | 1,758,233 | 940 | 828 |
| 9 | 385.94 | 0.18 | 157.31 | Tridecane | C13H28 | 603,935 | 922 | 821 |
| 10 | 1011.24 | 0.05 | 189.44 | Octadecane, 4-methyl- | C19H40 | 176,886 | 927 | 174 |
| 11 | 1085.77 | 0.0475 | 130.6396 | 1-Iodo-2-methylundecane | C12H25I | 156,952 | 934 | 136.5 |
| 12 | 1111.10 | 1.06 | 263.30 | Eicosane | C20H42 | 3,600,474 | 965 | 1270 |
| 13 | 1328.84 | 0.14 | 196.89 | Octacosane | C28H58 | 480,809 | 954 | 243 |
| 14 | 1225.79 | 0.62 | 239.27 | Tetracosane | C24H50 | 2,113,643 | 958 | 801 |
| 15 | 1098.44 | 0.16 | 216.14 | 5-Eicosene, (E)- | C20H40 | 531,752 | 842.50 | 353.50 |
| 16 | 1107.145 | 0.13 | 182.14295 | 3-Eicosene, (E)- | C20H40 | 432,411 | 954.5 | 232.5 |
| 17 | 743.364 | 0.197 | 206.1307 | 7-Hexadecene, (Z)- | C16H32 | 666,803 | 944 | 439 |
| Esters | ||||||||
| 18 | 421.96 | 0.03 | 150.07 | Benzeneacetic acid, methyl ester | C9H10O2 | 97,981 | 908 | 204 |
| 19 | 1061.66 | 0.0275 | 186.561 | 1,2-Benzenedicarboxylic acid, butyl 2-ethylhexyl ester | C20H30O4 | 87,747.5 | 22,613.625 | 480 |
| 20 | 1067.12 | 0.03 | 135.07 | Formic acid, 2-phenylethyl ester | C9H10O2 | 99,162 | 891 | 210 |
| 21 | 1080.37 | 0.02 | 292.20 | Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, methyl ester | C18H28O3 | 81,726 | 715 | 164 |
| 22 | 1061.69 | 0.03 | 187.06 | Dibutyl phthalate | C16H22O4 | 98,061 | 916 | 275 |
| 23 | 1059.04 | 0.30 | 219.63 | Ethyl 2-cyano-3-(4-methacryloyloxyphenyl)acrylate | C16H15NO4 | 1,011,529 | 866 | 1293 |
| Ketones | ||||||||
| 24 | 916.74 | 0.04 | 169.09 | 3-Methyl-1,4-diazabicyclo[4.3.0]nonan-2,5-dione, N-acetyl- | C10H14N2O3 | 143,741 | 758 | 493 |
| 25 | 951.08 | 0.04 | 154.07 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- | C7H10N2O2 | 145,827 | 848 | 459 |
| 26 | 1045.79 | 0.38 | 183.43 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl)- | C11H18N2O2 | 1,302,535 | 845 | 995 |
| 27 | 374.04 | 0.15 | 145.78 | 4-Piperidinone, 2,2,6,6-tetramethyl- | C9H17NO | 514,662 | 758 | 1406 |
| Other organic compounds | ||||||||
| 28 | 167.99 | 8.01 | 56.54 | Acetic acid, hydroxy- | C2H4O3 | 27,263,571 | 869 | 1676 |
| 29 | 273.27 | 0.01 | 121.09 | Pyridine, 2,4,6-trimethyl- | C8H11N | 6216 | 808 | 122 |
| 30 | 611.31 | 0.01 | 131.07 | Indole, 3-methyl- | C9H9N | 37,823 | 881 | 200 |
| 31 | 782.83 | 0.04 | 183.10 | Dodecanoic acid | C12H24O2 | 111,574 | 868 | 186 |
| 32 | 1087.75 | 0.04 | 228.20 | n-Hexadecanoic acid | C16H32O2 | 124,987 | 852 | 159 |
| 33 | 1215.02 | 0.10 | 211.11 | l-Leucyl-d-leucine | C12H24N2O3 | 312,139 | 717 | 322 |
| 34 | 1320.19 | 0.10 | 244.62 | Ergotaman-3′,6′,18-trione, 9,10-dihydro-12′-hydroxy-2′-methyl-5′-(phenylmethyl)-, (5′a,10a)- | C33H37N5O5 | 351,941 | 852 | 643 |
| No | R.T. (s) | Area% | Observed Ion m/z | Name | Formula | Area | Similarity | Peak S/N |
|---|---|---|---|---|---|---|---|---|
| Alcohols | ||||||||
| 1 | 966.5 | 0.11 | 146.8 | (3S,6S)-3-Butyl-6-methylpiperazine-2,5-dione | C9H16N2O2 | 913,414 | 723 | 559 |
| 2 | 697.2 | 0.03 | 155.1 | 1-Dodecanol | C12H26O | 285,215 | 919 | 140 |
| 3 | 623.9 | 0.03 | 155.1 | 1-Undecanol | C11H24O | 287,760 | 871 | 138 |
| 4 | 951.2 | 0.07 | 211.2 | 2,4-Di-tert-butylphenol | C14H22O | 536,471 | 802 | 366 |
| 5 | 763.4 | 0.09 | 127.6 | 1-Octen-4-ol | C8H16O | 270,020 | 790 | 401 |
| 6 | 520.0 | 0.05 | 128.0 | 2-Ethyl-1-hexanol | C8H18O | 143,985 | 839 | 192 |
| 7 | 786.7 | 0.03 | 138.2 | 2-Undecen-4-ol | C11H22O | 111,899 | 795 | 403 |
| 8 | 763.5 | 0.07 | 121.1 | 3-Mercapto-3-methylbutanol | C5H12OS | 286,892 | 789 | 623 |
| 9 | 974.8 | 0.03 | 126 | 4-Mercaptophenol | C6H6OS | 236,549 | 758 | 244 |
| 10 | 512.7 | 0.01 | 137.7 | Ethanol, 1-methoxy-, benzoate | C10H12O3 | 126,066 | 778 | 138 |
| 11 | 606.2 | 0.79 | 166.1 | Ethanol, 2-(2-butoxyethoxy)- | C8H18O3 | 6,801,240 | 886 | 17,274 |
| 12 | 646.6 | 0.26 | 141.4 | Ethanol, 2-(2-butoxyethoxy)-, acetate | C10H20O4 | 2,226,963 | 918 | 1609 |
| 13 | 281.0 | 0.10 | 117.2 | Ethanol, 2-(2-ethoxyethoxy)- | C6H14O3 | 650,282 | 833 | 365 |
| 14 | 942.1 | 0.04 | 133.6 | Ethanol, 2-[2-(ethenyloxy)ethoxy]- | C6H12O3 | 306,041 | 787 | 227 |
| 15 | 164.1 | 0.31 | 48.0 | Ethanol, 2,2-dichloro- | C2H4Cl2O | 967,422 | 947 | 142 |
| 16 | 269.7 | 8,42 | 32.0 | Methyl Alcohol | CH4O | 16,979,127 | 926 | 689 |
| 17 | 1161.2 | 0.04 | 139.8 | n-Tetracosanol-1 | C24H50O | 166,939 | 936 | 108 |
| 18 | 1393.3 | 0.04 | 368.3 | Phenol, 2,2′-methylenebis[6-(1,1-dimethylethyl)-4-ethyl- | C25H36O2 | 323,970 | 889 | 1769 |
| 19 | 738.2 | 0.02 | 206.2 | Phenol, 2,5-bis(1,1-dimethylethyl)- | C14H22O | 34,536 | 890 | 161 |
| 20 | 1246.6 | 0.17 | 228.1 | Phenol′, 4,4′-(1-methylethylidene)bis- | C15H16O2 | 1,403,098 | 726 | 378 |
| 21 | 371.7 | 0.08 | 122.1 | Phenylethyl Alcohol | C8H10O | 455,009 | 899 | 653 |
| Alkanes and Alkenes | ||||||||
| 22 | 1258.1 | 0.01 | 259.1 | 1,3-Dioxolane-2-heptanenitrile, a-methyl-d-oxo-2-phenyl- | C17H21NO3 | 65,288 | 747 | 150 |
| 23 | 988.2 | 0.48 | 124.4 | 1,5-Heptadiene, 3,4-dimethyl- | C9H16 | 4,164,767 | 869 | 1555 |
| 24 | 1206.7 | 0.04 | 144.6 | 1,3-Cyclopentadiene, 5-(trans-2-ethyl-3-methylcyclopropylidene)- | C11H14 | 332,887 | 723 | 520 |
| 25 | 877.1 | 0.02 | 169.2 | 1-Docosene | C22H44 | 186,631 | 874 | 195 |
| 26 | 296.7 | 0.05 | 100.6 | 2-Decene, 5-methyl-, (Z)- | C11H22 | 55,377 | 848 | 128 |
| 27 | 825.5 | 0.03 | 172.6 | 3-Eicosene, (E)- | C20H40 | 51,075 | 759 | 166 |
| 28 | 708.5 | 0.06 | 182.1 | 3-Tetradecene, (Z)- | C14H28 | 178,811 | 932 | 242 |
| 29 | 964.4 | 0.12 | 158.1 | 4-Heptafluorobutyryloxyhexadecane | C20H33F7O2 | 346,559 | 941 | 255 |
| 30 | 673.6 | 0.07 | 143.3 | 5-Esene, (E)- | C20H40 | 269,736 | 806 | 316 |
| 31 | 812.1 | 0.18 | 150.4 | 7-Hexadecene, (Z)- | C16H32 | 487,592 | 940 | 378 |
| 32 | 974.1 | 0.09 | 175.2 | 9-Eicosene, (E)- | C20H40 | 766,434 | 909 | 245 |
| 33 | 1057.7 | 1.34 | 237.9 | Eicosane | C20H42 | 2,954,435 | 959 | 1373 |
| 34 | 726.1 | 0.04 | 123.4 | Heptadecane, 2,6,10,14-tetramethyl- | C21H44 | 141,931 | 900 | 122 |
| 35 | 956.2 | 0.05 | 126.8 | Heptadecane, 2-methyl- | C18H38 | 161,042 | 929 | 160 |
| 36 | 1625.0 | 0.08 | 211.4 | Heptaethylene glycol | C14H30O8 | 209,475 | 930 | 166 |
| 37 | 1452.7 | 0.08 | 527.5 | Heptasiloxane, hexadecamethyl- | C16H48O6Si7 | 690,174 | 758 | 417 |
| 38 | 794.7 | 0.84 | 171.9 | Hexadecane | C16H34 | 1,956,218 | 943 | 1681 |
| 39 | 1486.3 | 0.07 | 159.4 | Hexaethylene glycol | C12H26O7 | 196,923 | 898 | 173 |
| 40 | 1348.6 | 0.21 | 182.9 | Octacosane | C28H58 | 509,942 | 953 | 264 |
| 41 | 931.3 | 0.06 | 156.5 | Octadecane, 4-methyl- | C19H40 | 135,195 | 898 | 164 |
| 42 | 1237.8 | 0.76 | 247.1 | Tetracosane | C24H50 | 2,390,307 | 955 | 855 |
| 43 | 548.8 | 0.28 | 153.0 | Tridecane | C13H28 | 712,424 | 918 | 798 |
| 44 | 868.7 | 0.04 | 127.1 | Tridecane, 4-methyl- | C14H30 | 116,341 | 907 | 130 |
| Esters | ||||||||
| 45 | 1005.1 | 0.06 | 143.4 | 1,2-Benzenedicarboxylic acid, butyl 2-ethylhexyl ester | C20H30O4 | 523,317 | 727 | 365 |
| 46 | 1092.4 | 0.03 | 150.0 | 1,2-Benzenedicarboxylic acid, dipropyl ester | C14H18O4 | 97,603 | 886 | 539 |
| 47 | 1546.4 | 0.97 | 294.5 | 1,2-Benzenedicarboxylic acid, decyl octyl ester | C26H42O4 | 715,862 | 893 | 152 |
| 48 | 1056.9 | 0.01 | 183.4 | 1H-Indole-3-ethanol, acetate (ester) | C12H13NO2 | 88,047 | 829 | 250 |
| 49 | 1377.6 | 0.01 | 195.0 | 2-Fluoro-3-trifluoromethylbenzoic acid, 3-methylbutyl-2 ester | C13H14F4O2 | 92,029 | 781 | 513 |
| 50 | 904.9 | 0.03 | 149.5 | 2-Propenoic acid, 2-methyl-, oxiranylmethyl ester | C7H10O3 | 107,925 | 792 | 425 |
| 51 | 668.6 | 0.04 | 126.8 | 2-Propenoic acid, tridecyl ester | C10H18N2O2 | 115,873 | 798 | 381 |
| 52 | 1308.8 | 0.01 | 273.5 | 2,2-diphenylpropionic acid, 2,2,2-trifluoroethyl ester | C17H15F3O2 | 101,730 | 743 | 173 |
| 53 | 266.8 | 0.18 | 86. | Acetic acid ethenyl ester | C4H6O2 | 437,551 | 975 | 312 |
| 54 | 754.1 | 0.01 | 194.1 | Benzoic acid, 4-ethoxy-, ethyl ester | C11H14O3 | 82,696 | 868 | 365 |
| 55 | 1563.6 | 0.04 | 180.0 | Carbonic acid, nonyl vinyl ester | C12H22O3 | 339,969 | 876 | 107 |
| 56 | 765.8 | 0.14 | 202.2 | Cyclopropanecarboxylic acid, 2-ethylhexyl ester | C12H22O2 | 1,233,173 | 811 | 1009 |
| 57 | 1132.1 | 0.05 | 154.1 | Di(1-methylcyclobutyl) ether | C10H18O | 370,096 | 814 | 213 |
| 58 | 819.8 | 0.01 | 177.7 | Diethyl Phthalate | C12H14O4 | 112,334 | 938 | 539 |
| 59 | 1404.6 | 0.06 | 204.6 | Diisooctyl phthalate | C24H38O4 | 67,422 | 860 | 340 |
| 60 | 1088.3 | 1.33 | 215.4 | DL-Alanine, N-methyl-N-(byt-3-yn-1-yloxycarbonyl)-, tetradecyl ester | C23H41NO4 | 11,535,230 | 890 | 817 |
| 61 | 892.0 | 0.08 | 186.5 | Hexanedioic acid, bis(2-methylpropyl) ester | C14H26O4 | 649,583 | 900 | 3117 |
| 62 | 1398.2 | 0.39 | 282.6 | l-Norvaline, n-propargyloxycarbonyl-, nonyl ester | C18H31NO4 | 1,742,412 | 909 | 232 |
| 63 | 1039.5 | 0.64 | 192.1 | L-Proline, N-valeryl-, decyl ester | C20H37NO3 | 5,550,110 | 788 | 850 |
| 64 | 1568.0 | 0.09 | 237.7 | Oxalic acid, allyl decyl ester | C15H26O4 | 809,220 | 907 | 186 |
| 65 | 1356.1 | 0.53 | 363.7 | Phosphoric acid, isodecyl diphenyl ester | C22H31O4P | 4,518,474 | 881 | 5661 |
| 66 | 1367.5 | 0.03 | 184.1 | Phosphoric acid, tris(2-ethylhexyl) ester | C24H51O4P | 211,680 | 804 | 951 |
| 67 | 1559.3 | 0.35 | 293.5 | Phthalic acid, 4-chloro-2-methylphenyl tetradecyl ester | C29H39ClO4 | 381,354 | 863 | 111 |
| 68 | 1544.1 | 0.86 | 294.2 | Phthalic acid, 7-methyloct-3-yn-5-yl undecyl ester | C28H42O4 | 771,078 | 861 | 152 |
| 69 | 1472.5 | 0.66 | 321.1 | Phthalic acid, 8-chlorooctyl decyl ester | C26H41ClO4 | 1,124,586 | 901 | 2078 |
| 70 | 841.5 | 0.07 | 192.1 | Propanoic acid, 2-methyl-, 2-phenylethyl ester | C12H16O2 | 564,174 | 734 | 532 |
| 71 | 0.00 | 0.001 | 515.4 | Propanoic acid, 3,3′-thiobis-, didodecyl ester | C30H58O4S | 456,850 | 748 | 487 |
| 72 | 1064.7 | 0.01 | 227.2 | Tridecanoic acid, methyl ester | C14H28O2 | 61,779 | 814 | 144 |
| 73 | 1065.2 | 0.01 | 206.6 | Undecanoic acid, methyl ester | C12H24O2 | 98,910 | 813 | 135 |
| Ketones | ||||||||
| 74 | 1166.8 | 0.04 | 225.1 | 1,3-Propanedione, 2-bromo-1,3-diphenyl- | C15H11BrO2 | 320,260 | 858 | 575 |
| 75 | 1043.6 | 0.01 | 140.1 | 1-(2-Thienyl)-1-propanone | C7H8OS | 133,221 | 731 | 172 |
| 76 | 921.0 | 0.16 | 205.1 | 2,2-Dimethyl-N-phenethylpropionamide | C13H19NO | 1,219,924.7 | 792.4 | 786.5 |
| 77 | 1233.1 | 0.0 | 204.1 | 2,5-Piperazinedione, 3-(phenylmethyl)- | C11H12N2O2 | 146,740 | 839 | 110 |
| 78 | 472.4 | 0.05 | 134.0 | 2-Coumaranone | C8H6O2 | 154,354 | 877 | 274 |
| 79 | 743.4 | 0.06 | 155.5 | 2-Dodecanone | C12H24O | 232,592 | 831 | 256 |
| 80 | 432.2 | 0.05 | 104.0 | 2-Pentanone, 4-hydroxy-4-methyl- | C6H12O2 | 123,821 | 804 | 337 |
| 81 | 381.7 | 0.03 | 143.1 | 4-Butoxy-2-butanone | C8H16O2 | 76,347 | 752 | 153 |
| 82 | 373.5 | 0.10 | 145.8 | 4-Piperidinone, 2,2,6,6-tetramethyl- | C9H17NO | 419,489 | 775 | 613 |
| 83 | 1018.1 | 0.31 | 154.1 | 5-Pyrrolidino-2-pyrrolidone | C8H14N2O | 1,355,861 | 709 | 477 |
| 84 | 1501.5 | 0.03 | 116.9 | Acetone, 1-[4-(dimethylaminoethoxy)phenyl]- | C13H19NO2 | 52,377 | 925 | 258 |
| 85 | 218.6 | 0.02 | 94.0 | Dimethyl sulfone | C2H6O2S | 63,528 | 889 | 472 |
| 86 | 188.0 | 0.02 | 78.0 | Dimethyl Sulfoxide | C2H6OS | 54,269 | 781 | 147 |
| 87 | 910.8 | 0.07 | 185.1 | Cyclopenta[c]quinolin-4-one, 1,2,3,5-tetrahydro- | C12H11NO | 110,533 | 807 | 574 |
| 88 | 1283.8 | 0.01 | 260.1 | Cyclopentanone, 2,5-bis(phenylmethylene)- | C19H16O | 33,095 | 808 | 182 |
| 89 | 1076.1 | 0.11 | 218.0 | Diphenyl sulfone | C12H10O2S | 285,983 | 818 | 1552 |
| 90 | 956.1 | 0.05 | 154.1 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- | C7H10N2O2 | 135,989 | 880 | 114 |
| 91 | 1048.4 | 2.08 | 203.6 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl)- | C11H18N2O2 | 5,818,832 | 819 | 2202 |
| Other organic compounds | ||||||||
| 92 | 857.6 | 0.01 | 161.1 | 3-Benzyl-5-chloro-1,2,3-triazole 1-oxide | C9H8ClN3O | 63,591 | 802 | 121 |
| 93 | 1112.1 | 0.02 | 241.1 | Metolachlor | C15H22ClNO2 | 199,899 | 775 | 1110 |
| 94 | 793.3 | 0.05 | 184.1 | 3-(2-Phenylethyl)pyridazine | C12H12N2 | 38,560 | 716 | 211 |
| 95 | 1121.6 | 0.10 | 475.0 | 3-Isopropoxy-1,1,1,7,7,7-hexamethyl-3,5,5-tris(trimethylsiloxy)tetrasiloxane | C18H52O7Si7 | 835,937 | 718 | 824 |
| 96 | 917.8 | 0.09 | 168.7 | 3-Methyl-1,4-diazabicyclo[4.3.0]nonan-2,5-dione, N-acetyl- | C10H14N2O3 | 208,480 | 790 | 309 |
| 97 | 224.7 | 0.37 | 190.5 | 4-(2-Acetoxyphenyl)-1-ethyl-3-methyl-5-(4-nitrophenyl)pyrazole | C20H19N3O4 | 3,198,062 | 999 | 332 |
| 98 | 1105.6 | 0.03 | 182.1 | 9H-Pyrido[3,4-b]indole, 1-methyl- | C12H10N2 | 60,640 | 878 | 306 |
| 99 | 1051.0 | 0.05 | 235.2 | Acetamide, 2-chloro-N-(ethoxymethyl)-N-(2-ethyl-6-methylphenyl)- | C14H20ClNO2 | 455,685 | 760 | 507 |
| 100 | 177.0 | 8.87 | 66.5 | Acetic acid, hydroxy- | C2H4O3 | 39,385,471 | 788 | 2051 |
| 101 | 267.8 | 0.11 | 109.5 | Acetic anhydride | C4H6O3 | 322,969 | 995 | 273 |
| 102 | 1215.3 | 0.08 | 222.1 | Benzene, (1,2-dicyclopropyl-2-phenylethyl)- | C20H22 | 696,698 | 732 | 371 |
| 103 | 865.4 | 0.01 | 132.1 | Benzene, (1-azido-1-methylethyl)- | C9H11N3 | 83,422 | 761 | 165 |
| 104 | 621.3 | 0.01 | 135.1 | Benzeneacetamide | C8H9NO | 100,313 | 846 | 217 |
| 105 | 519.7 | 0.04 | 146.1 | Benzeneethanamine, N-(1-methylethylidene)- | C11H15N | 125,968 | 879 | 135 |
| 106 | 651.1 | 0.09 | 165.5 | Benzeneethanamine, N-(3-methylbutylidene)- | C13H19N | 254,728 | 847 | 133 |
| 107 | 652.6 | 0.04 | 138.1 | Benzeneethanol, 4-hydroxy- | C8H10O2 | 309,979 | 876 | 356 |
| 108 | 962.9 | 0.04 | 212.1 | Benzyl Benzoate | C14H12O2 | 361,122 | 873 | 444 |
| 109 | 603.3 | 0.06 | 134.1 | Bicyclo[3.1.0]hex-2-ene, 4-methylene-1-(1-methylethyl)- | C10H14 | 111,702 | 756 | 259 |
| 110 | 1309.0 | 0.02 | 273.6 | Bifenthrin | C23H22ClF3O2 | 135,178 | 725 | 284 |
| 111 | 1406.2 | 0.76 | 390.6 | Bis(2-ethylhexyl) phthalate | C24H38O4 | 6,611,090 | 900 | 5447 |
| 112 | 1416.0 | 0.03 | 315.1 | Bumetrizole | C17H18ClN3O | 302,918 | 943 | 1675 |
| 113 | 518.3 | 0.05 | 156.1 | Butanamide, N-hexyl- | C10H21NO | 465,622 | 724 | 421 |
| 114 | 181.5 | 2.30 | 97.0 | Butanoic acid, 3-methyl- | C5H10O2 | 1,955,281 | 856 | 632 |
| 115 | 743.7 | 0.09 | 220.2 | Butylated Hydroxytoluene | C15H24O | 802,322 | 931 | 3077 |
| 116 | 964.6 | 0.10 | 137.4 | Cyclo-(glycyl-l-leucyl) | C8H14N2O2 | 198,566 | 760 | 171 |
| 117 | 1536.4 | 0.04 | 86.1 | Cyclobutane, methoxy- | C5H10O | 164,364 | 891 | 431 |
| 118 | 877.9 | 0.09 | 437.0 | Cyclooctasiloxane, hexadecamethyl- | C16H48O8Si8 | 791,787 | 719 | 1152 |
| 119 | 1361.3 | 0.17 | 287.5 | Di-n-octyl phenyl phosphate | C22H39O4P | 1,439,556 | 733 | 5036 |
| 120 | 961.9 | 0.27 | 168.8 | dl-Alanyl-l-leucine | C9H18N2O3 | 2,322,399 | 853 | 247 |
| 121 | 1000.9 | 0.02 | 201.7 | Dodecanoic acid, 2-methyl- | C13H26O2 | 210,456 | 857 | 317 |
| 122 | 860.5 | 0.03 | 172.6 | Dodecanoic acid | C12H24O2 | 113,150 | 832 | 158 |
| 123 | 898.2 | 0.04 | 253.3 | Dodecyl acrylate | C15H28O2 | 347,139 | 939 | 273 |
| 124 | 1330.3 | 2.56 | 291.3 | Ergotaman-3′,6′,18-trione, 9,10-dihydro-12′-hydroxy-2′-methyl-5′-(phenylmethyl)-, (5′α,10a)- | C33H37N5O5 | 7,364,698 | 870 | 2348 |
| 125 | 340.0 | 0.04 | 103.6 | Glycyl-dl-norvaline | C7H14N2O3 | 362,257 | 773 | 431 |
| 126 | 1665.0 | 0.02 | 432.1 | Hexasiloxane, tetradecamethyl- | C14H42O5Si6 | 170,683 | 750 | 131 |
| 127 | 1648.3 | 0.03 | 342.3 | Indeno[1,2-b]pyridine, 7-methyl-5-(2,2,6,6-tetramethylpiperid-4-ylimino)- | C22H27N3 | 270,671 | 866 | 1478 |
| 128 | 522.0 | 0.01 | 117.1 | Indole | C8H7N | 63,439 | 854 | 329 |
| 129 | 484.7 | 0.25 | 146.1 | Isobutyramide, N-(3-methylbutyl)- | C9H19NO | 2,176,992 | 765 | 1702 |
| 130 | 708.8 | 0.00 | 146.1 | c, 1,2,3,4-tetrahydro-1,8-dimethyl- | C12H16 | 14,327 | 729 | 124 |
| 131 | 1148.5 | 0.05 | 227.5 | n-Hexadecanoic acid | C16H32O2 | 258,286 | 825 | 177 |
| 132 | 358.7 | 0.03 | 106.1 | Nonanal | C9H18O | 110,278 | 815 | 162 |
| 133 | 854.5 | 0.02 | 167.1 | Octanamide, N,N-dimethyl- | C10H21NO | 162,285 | 796 | 836 |
| 134 | 1352.4 | 0.06 | 252.0 | Octicizer | C20H27O4P | 46,610 | 810 | 122 |
| 135 | 1332.4 | 0.37 | 247.6 | Oxamide, N-(3-m′thoxypropyl)-N′-cycloheptylidenamino- | C13H23N3O3 | 3,237,962 | 953 | 828 |
| 136 | 163.1 | 1.67 | 32.0 | Oxygen | O2 | 5,306,645 | 957 | 488 |
| 137 | 339.0 | 0.05 | 103.6 | Pentanoic acid | C5H10O2 | 424,336 | 741 | 250 |
| 138 | 832.1 | 0.05 | 191.1 | Phenylacetamide, N-isobutyl- | C12H17NO | 387,577 | 819 | 640 |
| 139 | 922.9 | 0.10 | 209.1 | Phenylacetamide, N-pentyl- | C13H19NO | 788,730 | 836 | 282 |
| 140 | 263.5 | 0.09 | 182.3 | Phosphonic acid, (p-hydroxyphenyl)- | C6H7O4P | 817,511 | 814 | 3495 |
| 141 | 1347.1 | 0.03 | 260.1 | terphenyl, 4,4″-diamine | C18H16N2 | 77,775 | 817 | 414 |
| 142 | 793.4 | 0.03 | 184.1 | Pyridine, 3-(4-tolylamino)- | C12H12N2 | 101,812 | 729 | 552 |
| 143 | 1144.4 | 0.02 | 184.8 | Quinoline, 2-(2-methylpropyl)- | C13H15N | 180,409 | 750 | 238 |
| 144 | 890.7 | 0.02 | 191.5 | ß-Phenylethyl butyrate | C12H16O2 | 225,091 | 759 | 162 |
| 145 | 1344.8 | 0.12 | 326.1 | Triphenyl phosphate | C18H15O4P | 1,059,663 | 917 | 648 |
| 146 | 938.5 | 0.06 | 1878 | Undecanoic acid | C11H22O2 | 351,308 | 852 | 235 |
| 147 | 537.7 | 0.34 | 16.8 | Valeramide, N-hexyl- | C11H23NO | 2,316,095 | 730 | 3538 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nchabeleng, M.M.; Fonkui, T.Y.; Ezekiel, G. The Indiscriminate Chemical Makeup of Secondary Metabolites Derived from Endophytes Harvested from Aloe barbadensis Miller in South Africa’s Limpopo Region. Molecules 2024, 29, 1297. https://doi.org/10.3390/molecules29061297
Nchabeleng MM, Fonkui TY, Ezekiel G. The Indiscriminate Chemical Makeup of Secondary Metabolites Derived from Endophytes Harvested from Aloe barbadensis Miller in South Africa’s Limpopo Region. Molecules. 2024; 29(6):1297. https://doi.org/10.3390/molecules29061297
Chicago/Turabian StyleNchabeleng, Mpho Mamphoka, Thierry Youmbi Fonkui, and Green Ezekiel. 2024. "The Indiscriminate Chemical Makeup of Secondary Metabolites Derived from Endophytes Harvested from Aloe barbadensis Miller in South Africa’s Limpopo Region" Molecules 29, no. 6: 1297. https://doi.org/10.3390/molecules29061297
APA StyleNchabeleng, M. M., Fonkui, T. Y., & Ezekiel, G. (2024). The Indiscriminate Chemical Makeup of Secondary Metabolites Derived from Endophytes Harvested from Aloe barbadensis Miller in South Africa’s Limpopo Region. Molecules, 29(6), 1297. https://doi.org/10.3390/molecules29061297






