Identification and Biological Evaluation of a Novel Small-Molecule Inhibitor of Ricin Toxin
Abstract
:1. Introduction
2. Results
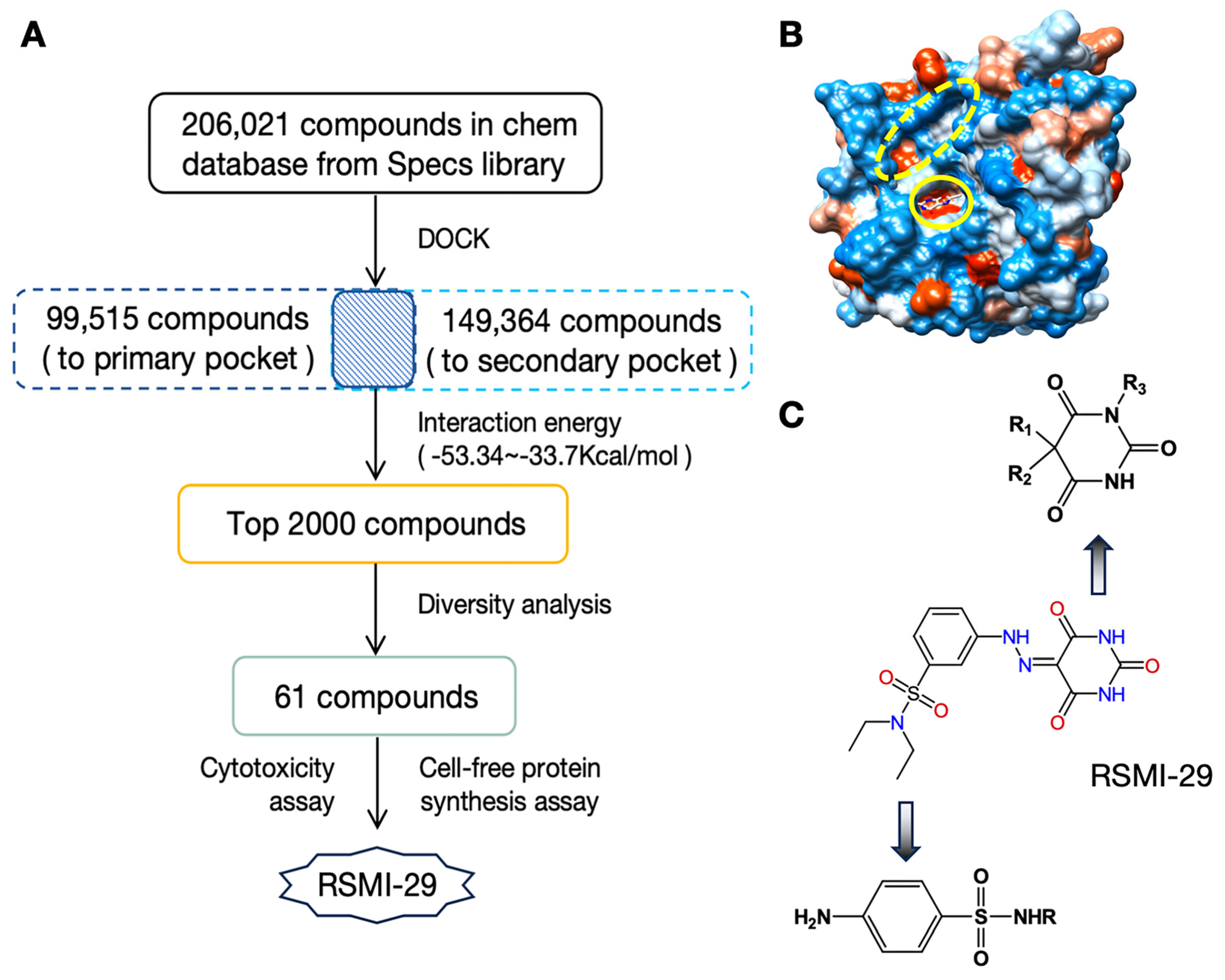
2.1. Discover Potential RTA Inhibitors by Virtual Screening
2.2. RSMI-29 Inhibits the Cytotoxicity of RTA/Ricin
2.3. RSMI-29 Inhibits the Biological Activity of RTA/Ricin In Vitro


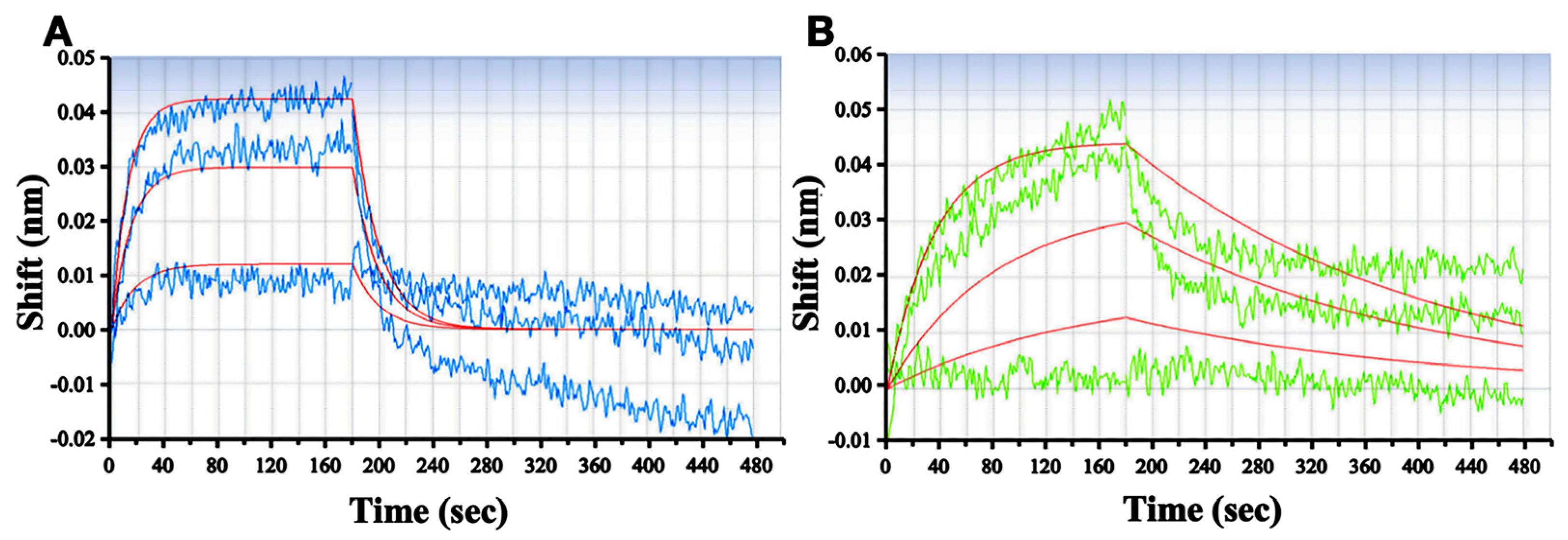
2.4. RSMI-29 Directly Binds to RTA
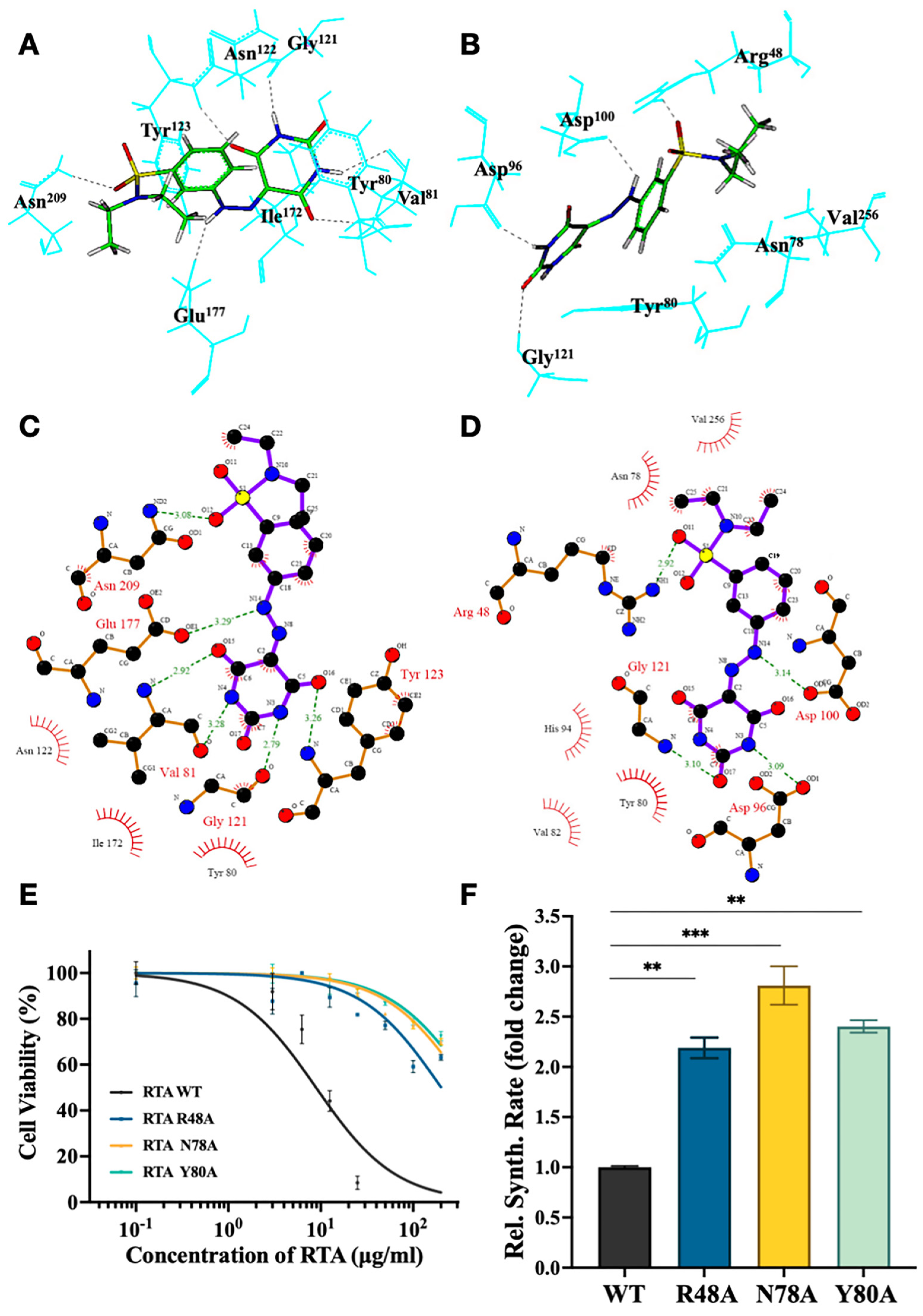
2.5. Identify the Important Amino Acid Residues of RSMI-29 Interacting with RTA
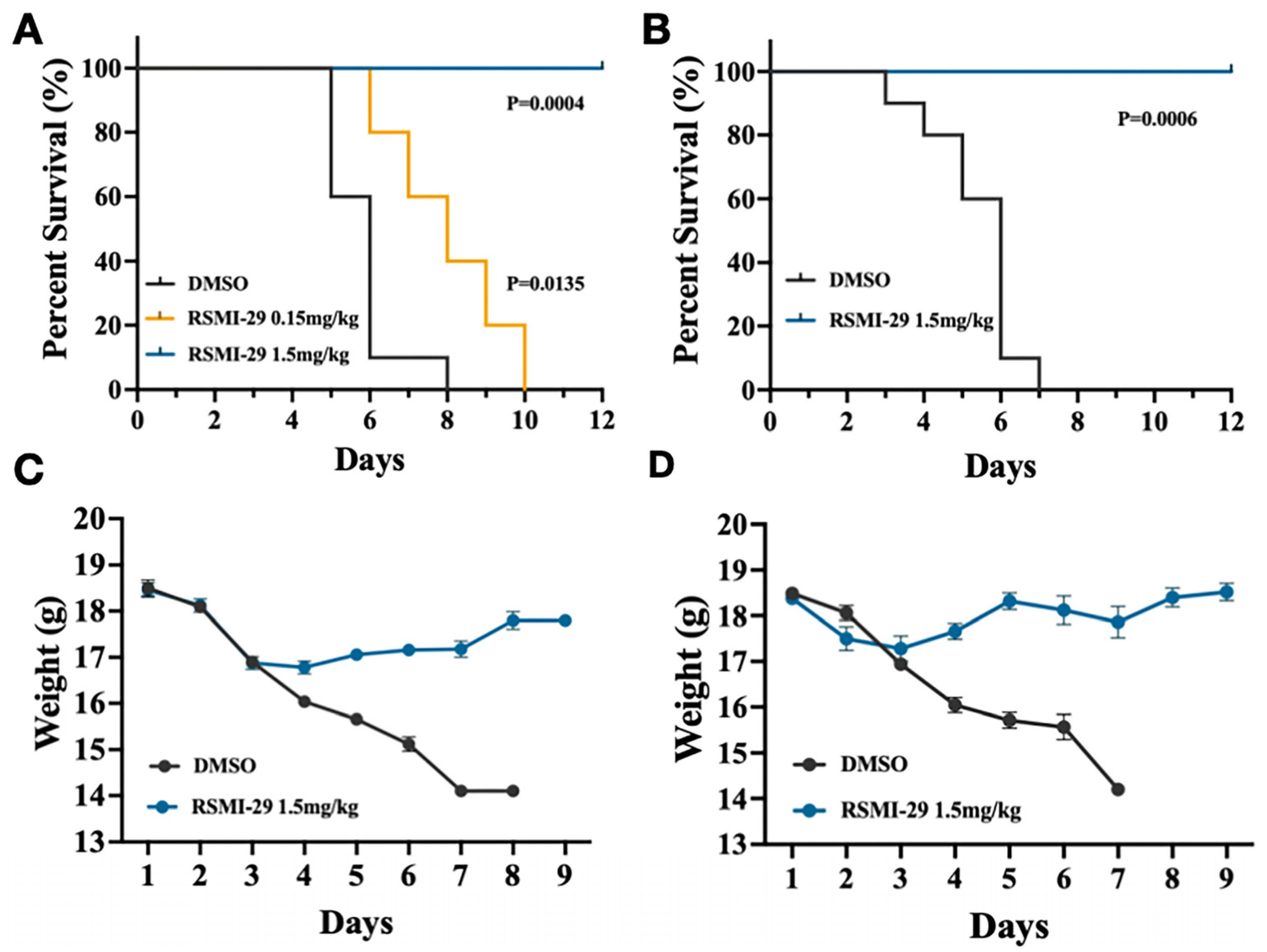
2.6. RSMI-29 Protects Mice against RICIN Intoxication
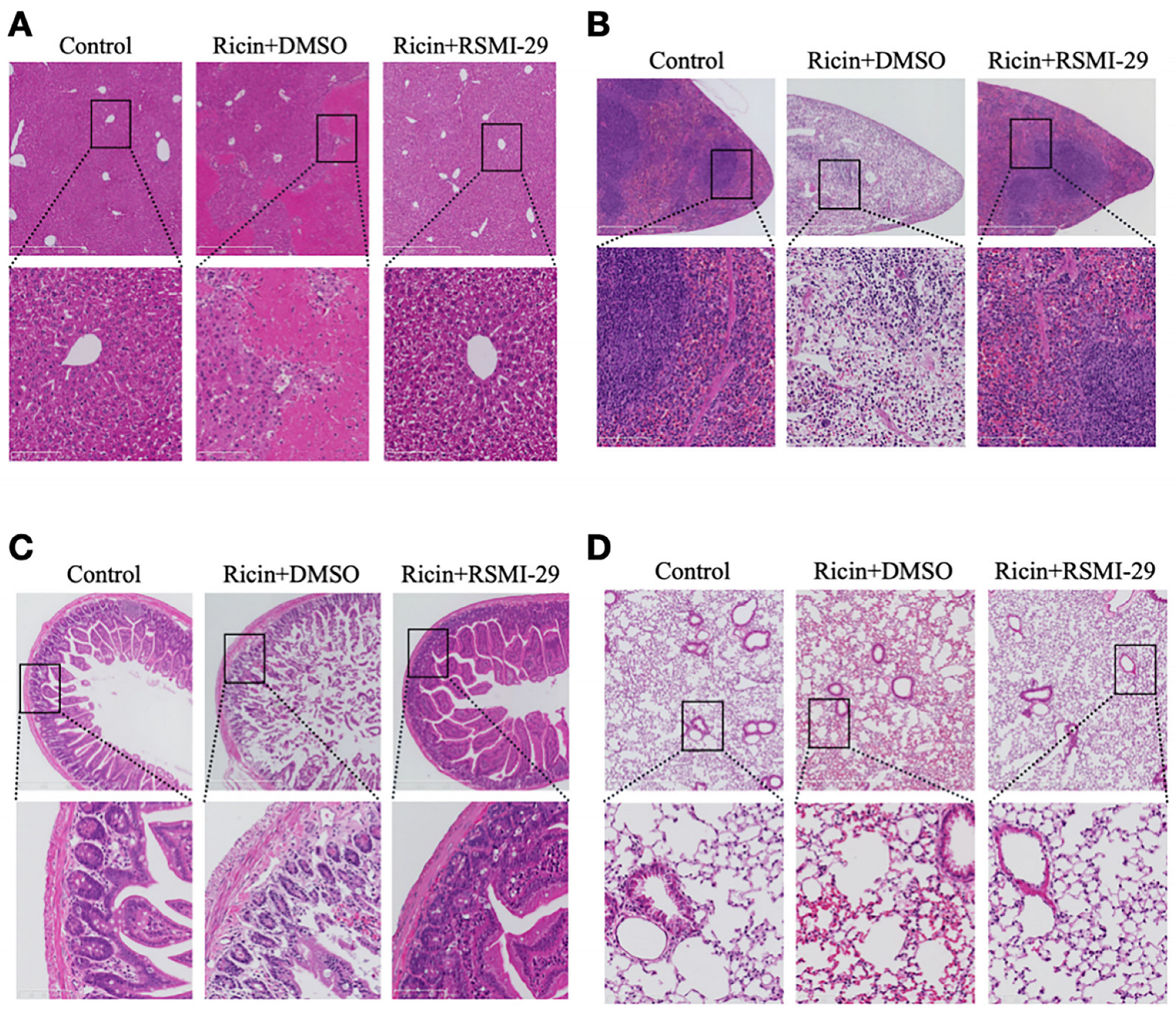
2.7. RSMI-29 Reduces Ricin-Induced Liver, Spleen, Intestine, and Lung Injury in Mice
3. Discussion
4. Materials and Methods
4.1. Chemical and Reagents
4.2. Cell Line and Culture Condition
4.3. Animals Feeding
4.4. Molecular Docking
4.5. Expression and Purification of RTA and Its Mutants
4.6. Biolayer Interferometry (BLI)
4.7. Cell-Free Luciferase Translation Assay
4.8. Cytotoxicity Assay
4.9. Depurination Assay
4.10. In Vivo Experiments
4.11. Histological Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sowa-Rogozińska, N.; Sominka, H.; Nowakowska-Gołacka, J.; Sandvig, K.; Słomińska Wojewódzka, M. Intracellular Transport and Cytotoxicity of the Protein Toxin Ricin. Toxins 2019, 11, 350. [Google Scholar] [CrossRef]
- Brunka, Z.; Ryl, J.; Brushtulli, P.; Gromala, D.; Walczak, G.; Zięba, S.; Pieśniak, D.; Anand, J.S.; Wiergowski, M. Selected Political Criminal Poisonings in the Years 1978–2020: Detection and Treatment. Toxics 2022, 10, 468. [Google Scholar] [CrossRef] [PubMed]
- Janik, E.; Ceremuga, M.; Saluk-Bijak, J.; Bijak, M. Biological Toxins as the Potential Tools for Bioterrorism. Int. J. Mol. Sci. 2019, 20, 1181. [Google Scholar] [CrossRef] [PubMed]
- Polito, L.; Bortolotti, M.; Battelli, M.G.; Calafato, G.; Bolognesi, A. Ricin: An Ancient Story for a Timeless Plant Toxin. Toxins 2019, 11, 324. [Google Scholar] [CrossRef]
- Abbes, M.; Montana, M.; Curti, C.; Vanelle, P. Ricin poisoning: A review on contamination source, diagnosis, treatment, prevention and reporting of ricin poisoning. Toxicon 2021, 195, 86–92. [Google Scholar] [CrossRef]
- Franke, H.; Scholl, R.; Aigner, A. Ricin and Ricinus communis in pharmacology and toxicology-from ancient use and “Papyrus Ebers” to modern perspectives and “poisonous plant of the year 2018”. Naunyn Schmiedebergs Arch. Pharmacol. 2019, 392, 1181–1208. [Google Scholar] [CrossRef] [PubMed]
- Spooner, R.A.; Lord, J.M. Ricin trafficking in cells. Toxins 2015, 7, 49–65. [Google Scholar] [CrossRef]
- Walsh, M.J.; Dodd, J.E.; Hautbergue, G.M. Ribosome-inactivating proteins: Potent poisons and molecular tools. Virulence 2015, 4, 774–784. [Google Scholar] [CrossRef]
- Fang, E.F.; Ng, T.B.; Shaw, P.C.; Wong, R.N. Recent progress in medicinal investigations on trichosanthin and other ribosome inactivating proteins from the plant genus Trichosanthes. Curr. Med. Chem. 2011, 18, 4410–4417. [Google Scholar] [CrossRef]
- Gaylord, S.T.; Dinh, T.L.; Goldman, E.R.; Anderson, G.P.; Ngan, K.C.; Walt, D.R. Ultrasensitive Detection of Ricin Toxin in Multiple Sample Matrixes Using Single-Domain Antibodies. Anal. Chem. 2015, 87, 6570–6577. [Google Scholar] [CrossRef]
- Yu, H.; Li, S.; Xu, N.; Liu, W. Ricin toxin and its neutralizing antibodies: A review. Toxicon 2022, 214, 47–53. [Google Scholar] [CrossRef]
- Sarkes, D.A.; Hurley, M.M.; Stratis-Cullum, D.N. Unraveling the Roots of Selectivity of Peptide Affinity Reagents for Structurally Similar Ribosomal Inactivating Protein Derivatives. Molecules 2016, 21, 1504. [Google Scholar] [CrossRef]
- Rasetti-Escargueil, C.; Avril, A. Medical Countermeasures against Ricin Intoxication. Toxins 2023, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Hollis, T.; Svinth, M.; Day, P.; Monzingo, A.F.; Milne, G.W.; Robertus, J.D. Structure-based identification of a ricin inhibitor. J. Mol. Biol. 1997, 266, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Pruet, J.M.; Jasheway, K.R.; Manzano, L.A.; Bai, Y.; Anslyn, E.V.; Robertus, J.D. 7-Substituted pterins provide a new direction for ricin A chain inhibitors. Eur. J. Med. Chem. 2011, 46, 3608–3615. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Pruet, J.M.; Manzano, L.A.; Jasheway, K.; Monzingo, A.F.; Wiget, P.A.; Kamat, I.; Anslyn, E.V.; Robertus, J.D. Peptide-conjugated pterins as inhibitors of ricin toxin A. J. Med. Chem. 2013, 56, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Pruet, J.M.; Saito, R.; Manzano, L.A.; Jasheway, K.R.; Wiget, P.A.; Kamat, I.; Anslyn, E.V.; Robertus, J.D. Optimized 5-membered heterocycle-linked pterins for the inhibition of Ricin Toxin A. ACS Med. Chem. Lett. 2012, 3, 588–591. [Google Scholar] [CrossRef]
- Wahome, P.G.; Robertus, J.D.; Mantis, N.J. Small-molecule inhibitors of ricin and Shiga toxins. Curr. Top. Microbiol. Immunol. 2012, 357, 179–207. [Google Scholar]
- França, T.C.C.; Botelho, F.D.; LaPlante, S.R. Theoretical Investigation of Repurposed Drugs Potentially Capable of Binding to the Catalytic Site and the Secondary Binding Pocket of Subunit A of Ricin. ACS Omega 2022, 7, 32805–32815. [Google Scholar] [CrossRef]
- Huang, N.; Nagarsekar, A.; Xia, G.; Hayashi, J.; MacKerell, A.D., Jr. Identification of non-phosphate-containing small molecular weight inhibitors of the tyrosine kinase p56 Lck SH2 domain via in silico screening against the pY + 3 binding site. J. Med. Chem. 2004, 47, 3502–3511. [Google Scholar] [CrossRef]
- Peng, H.; Huang, N.; Qi, J.; Xie, P.; Xu, C.; Wang, J.; Yang, C. Identification of novel inhibitors of BCR-ABL tyrosine kinase via virtual screening. Bioorg. Med. Chem. Lett. 2003, 13, 3693–3699. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.; Siva Prasad, C.V. Ligand based virtual screening to find novel inhibitors against plant toxin Ricin by using the ZINC database. Bioinformation 2011, 7, 46–51. [Google Scholar] [CrossRef]
- Cong, D.; Li, Y.; Ludford, P.T.; Tor, Y. Isomorphic Fluorescent Nucleosides Facilitate Real-Time Monitoring of RNA Depurination by Ribosome Inactivating Proteins. Chemistry 2022, 28, e202200994. [Google Scholar] [CrossRef]
- Kim, Y.; Robertus, J.D. Analysis of several key active site residues of ricin A chain by mutagenesis and X-ray crystallography. Protein Eng. 1992, 5, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Ready, M.P.; Kim, Y.; Robertus, J.D. Site-directed mutagenesis of ricin A-chain and implications for the mechanism of action. Proteins 1991, 10, 270–278. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 2015, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Sapoznikov, A.; Gal, Y.; Evgy, Y.; Aftalion, M.; Katalan, S.; Sabo, T.; Kronman, C.; Falach, R. Intramuscular Exposure to a Lethal Dose of Ricin Toxin Leads to Endothelial Glycocalyx Shedding and Microvascular Flow Abnormality in Mice and Swine. Int. J. Mol. Sci. 2021, 22, 12345. [Google Scholar] [CrossRef]
- Roy, C.J.; Song, K.; Sivasubramani, S.K.; Gardner, D.J.; Pincus, S.H. Animal models of ricin toxicosis. Curr. Top. Microbiol. Immunol. 2012, 357, 243–257. [Google Scholar]
- Sapoznikov, A.; Rosner, A.; Falach, R.; Gal, Y.; Aftalion, M.; Evgy, Y.; Israeli, O.; Sabo, T.; Kronman, C. Intramuscular Ricin Poisoning of Mice Leads to Widespread Damage in the Heart, Spleen, and Bone Marrow. Toxins 2019, 16, 344. [Google Scholar] [CrossRef]
- Roy, C.J.; Ehrbar, D.; Van Slyke, G.; Doering, J.; Didier, P.J.; Doyle-Meyers, L.; Donini, O.; Vitetta, E.S.; Mantis, N.J. Serum antibody profiling identifies vaccine-induced correlates of protection against aerosolized ricin toxin in rhesus macaques. NPJ Vaccines 2022, 7, 164. [Google Scholar] [CrossRef]
- Montgomery, V.A.; Lindsey, C.Y.; Smith, L.A.; Webb, R.P. Development of an o-pthalaldehyde (OPA) assay to measure protein content in Ricin Vaccine E. coli (RVEc™). Vaccine 2021, 39, 564–570. [Google Scholar] [CrossRef]
- Botelho, F.D.; Franca, T.C.C.; LaPlante, S.R. The Search for Antidotes Against Ricin. Mini Rev. Med. Chem. 2024. epub ahead of print. [Google Scholar] [CrossRef]
- Vance, D.J.; Rudolph, M.J.; Davis, S.A.; Mantis, N.J. Structural Basis of Antibody-Mediated Inhibition of Ricin Toxin Attachment to Host Cells. Biochemistry 2023, 62, 3181–3187. [Google Scholar] [CrossRef]
- Zhao, X.; Li, H.; Li, J.; Liu, K.; Wang, B.; Wang, Y.; Li, X.; Zhong, W. Novel small molecule retrograde transport blocker confers post-exposure protection against ricin intoxication. Acta Pharm. Sin. B 2020, 10, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Jasheway, K.; Pruet, J.; Anslyn, E.V.; Robertus, J.D. Structure-based design of ricin inhibitors. Toxins 2011, 3, 1233–1248. [Google Scholar] [CrossRef]
- Goto, M.; Sakamoto, N.; Higashi, S.; Kawata, R.; Nagatsu, K.; Saito, R. Crystal structure of ricin toxin A chain complexed with a highly potent pterin-based small-molecular inhibitor. J. Enzym. Inhib. Med. Chem. 2023, 38, 2219038. [Google Scholar] [CrossRef]
- Chaves, E.J.F.; Gomes da Cruz, L.E.; Padilha, I.Q.M.; Silveira, C.H.; Araujo, D.A.M.; Rocha, G.B. Discovery of RTA ricin subunit inhibitors: A computational study using PM7 quantum chemical method and steered molecular dynamics. J. Biomol. Struct. Dyn. 2022, 40, 5427–5445. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.B.; Suresh, M.X. A computational perspective of molecular interactions through virtual screening, pharmacokinetic and dynamic prediction on ribosome toxin A chain and inhibitors of Ricinus communis. Pharmacogn. Res. 2012, 4, 2–10. [Google Scholar] [CrossRef]
- Bai, Y.; Watt, B.; Wahome, P.G.; Mantis, N.J.; Robertus, J.D. Identification of new classes of ricin toxin inhibitors by virtual screening. Toxicon 2010, 56, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Capuzzi, S.J.; Muratov, E.N.; Tropsha, A. Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay INterference CompoundS. J. Chem. Inf. Model. 2017, 57, 417–427. [Google Scholar] [CrossRef]
- Phatak, P.; Chauhan, V.; Dhaked, R.K.; Pathak, U.; Saxena, N. E-N-(2-acetyl-phenyl)-3-phenyl-acrylamide targets abrin and ricin toxicity: Hitting two toxins with one stone. Biomed. Pharmacother. 2021, 143, 112134. [Google Scholar] [CrossRef]
- Sapoznikov, A.; Gal, Y.; Alcalay, R.; Evgy, Y.; Sabo, T.; Kronman, C.; Falach, R. Characterization of Lung Injury following Abrin Pulmonary Intoxication in Mice: Comparison to Ricin Poisoning. Toxins 2022, 14, 614. [Google Scholar] [CrossRef]
- Moustakas, D.T.; Lang, P.T.; Pegg, S.; Pettersen, E.; Kuntz, I.D.; Brooijmans, N.; Rizzo, R.C. Development and validation of a modular, extensible docking program: DOCK 5. J. Comput. Aided Mol. Des. 2006, 20, 601–619. [Google Scholar] [CrossRef]
- Miteva, M.A.; Lee, W.H.; Montes, M.O.; Villoutreix, B.O. Fast structure-based virtual ligand screening combining FRED, DOCK, and Surflex. J. Med. Chem. 2005, 48, 6012–6022. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Hu, M.R.; Guo, J.W.; Feng, J.N.; Shen, B.F. Fusion expression and purification of recombinant ricin A-chain. Chin. J. Cell. Mol. Immunol. 2005, 21, 137–140. (In Chinese) [Google Scholar]
- Xu, R.; Li, D.; Peng, J.; Fang, J.; Zhang, L.; Liu, L. Cloning, expression and antioxidant activity of a novel collagen from Pelodiscus sinensis. World J. Microbiol. Biotechnol. 2016, 32, 100. [Google Scholar] [CrossRef]
- Dong, N.; Luo, L.; Wu, J.; Jia, P.; Li, Q.; Wang, Y.; Gao, Z.; Peng, H.; Lv, M.; Huang, C.; et al. Monoclonal antibody, mAb 4C13, an effective detoxicant antibody against ricin poisoning. Vaccine 2015, 33, 3836–3842. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Z.; Piao, C.; Zhang, Z.; Kong, C.; Yin, L.; Liu, X. Flavokawain A is a natural inhibitor of PRMT5 in bladder cancer. J. Exp. Clin. Cancer Res. 2022, 41, 293. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, X.P.; Kahn, J.N.; Tumer, N.E. Functional Assays for Measuring the Catalytic Activity of Ribosome Inactivating Proteins. Toxins 2018, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Stechmann, B.; Bai, S.K.; Gobbo, E.; Lopez, R.; Merer, G.; Pinchard, S.; Panigai, L.; Tenza, D.; Raposo, G.; Beaumelle, B.; et al. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 2010, 141, 231–242. [Google Scholar] [CrossRef]
- Chen, Q.; Han, H.; Lin, F.; Yang, L.; Feng, L.; Lai, X.; Wen, Z.; Yang, M.; Wang, C.; Ma, Y.; et al. Novel shikonin derivatives suppress cell proliferation, migration and induce apoptosis in human triple-negative breast cancer cells via regulating PDK1/PDHC axis. Life Sci. 2022, 310, 121077. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Harijan, R.K.; Cao, B.; Kahn, J.N.; Pierce, M.; Tsymbal, A.M.; Roberge, J.Y.; Augeri, D.; Tumer, N.E. Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design. J. Med. Chem. 2021, 64, 15334–15348. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fang, B.; Huang, Y.; Tao, S.; Sun, B.; Guan, S.; Jin, Y. Epigallocatechin-3-gallate protects against 1,3-dichloro-2-propanol-induced lipid accumulation in C57BL/6J mice. Life Sci. 2018, 209, 324–331. [Google Scholar] [CrossRef] [PubMed]
| Compound | KD (M) | Kon (1/Ms) | Koff (1/s) |
|---|---|---|---|
| PTA | 8.68 × 10−6 | 5.92 × 103 | 5.14 × 10−2 |
| RSMI-29 | 9.23 × 10−7 | 8.32 × 103 | 7.68 × 10−3 |
| Forward Primers (5′-3′) | Reverse Primers (5′-3′) | |
|---|---|---|
| RTA R48A | GTTGCCAAACGCAGTTGGTTTGCCTATAAACC | ACCAACTGCGTTTGGCAACACTGGTATTTC |
| RTA N78A | GGATGTCACCGCTGCATATGTGGTCGGCTAC | CATATGCAGCGGTGA CATCCAGGGCTAATG |
| RTA Y80A | CACCAATGCAGCTGTGG TCGGCTACC | AGCCGACCACATATG CATTGGTGACATCCA |
| Human depurinated rRNA | TGCCATGGTAATCCTGCT CAGTA | TCTGAACCTGCGGTT CCACA |
| Human 28S rRNA | GATGTCGGCTCTTCCTAT CATTGT | CCAGCTCACGTTCCC TATTAGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Wei, A.; Cao, X.; Wang, Z.; Wan, H.; Wang, B.; Peng, H. Identification and Biological Evaluation of a Novel Small-Molecule Inhibitor of Ricin Toxin. Molecules 2024, 29, 1435. https://doi.org/10.3390/molecules29071435
Yang X, Wei A, Cao X, Wang Z, Wan H, Wang B, Peng H. Identification and Biological Evaluation of a Novel Small-Molecule Inhibitor of Ricin Toxin. Molecules. 2024; 29(7):1435. https://doi.org/10.3390/molecules29071435
Chicago/Turabian StyleYang, Xinran, Aili Wei, Xiyuan Cao, Zicheng Wang, Hongzhi Wan, Bo Wang, and Hui Peng. 2024. "Identification and Biological Evaluation of a Novel Small-Molecule Inhibitor of Ricin Toxin" Molecules 29, no. 7: 1435. https://doi.org/10.3390/molecules29071435






