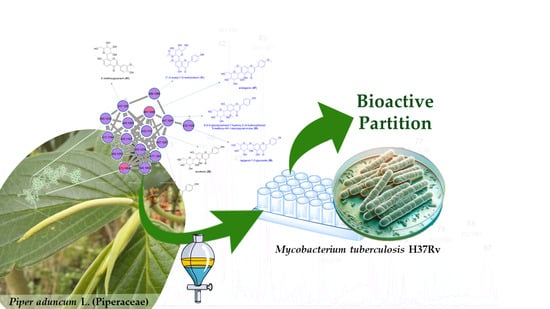
UHPLC-HRMS/MS Chemical Fingerprinting of the Bioactive Partition from Cultivated Piper aduncum L.
Abstract
:1. Introduction
2. Results and Discussion
2.1. Activity against Mycobacterium tuberculosis H37Rv
2.2. Chemical Composition Analysis of the Bioactive Partition
- (a)
- The monoterpenic lactone loliolide (24, m/z 197.1174, [M+H]+), a major constituent of SFR3, with seven fragment ions matching the GNPS library spectrum, previously described in the species Piper boehmeriifolium (Miq.) Wall. ex C.DC. [36];
- (b)
- The flavonoids wogonin (69, m/z 285.0758, [M+H]+) in SFR5, along with two glycosylated flavonoids, isoswertisin (32, m/z 447.1281, [M+H]+) and 7-(β-D-glucopyranosyloxy)-5-methoxy-2-(4-methoxyphenyl)-4H-1-benzopyran-4-one (42, m/z 461.1443, [M+H]+), with the latter being predominant in SFR5. These flavonoids did not cluster into the corresponding molecular families due to the low-intensity signals in the MS2 spectrum, preventing the calculation of similarity between spectra. The presence of fragment ions was observed in the raw data, allowing for the manual annotation of substances based on the similarity with MS2 spectra from libraries;
- (c)
- Two chalcones, 2,6-dihydroxy-4-methoxydihydrochalcone (57, m/z 273.1124, [M+H]+) and 2,4-dihydroxy-6-methoxydihydrochalcone (72, m/z 273.1118, [M+H]+), were identified in the SFR4 and SFR5 fractions, both with eight ions matching the spectra in the GNPS library. The analog of 57, 2,6-dihydroxy-4-methoxychalcone was previously isolated from P. aduncum by our group and exhibited great leishmanicidal activity [37,38];
- (d)
- Esculetin (2, m/z 179.0343, [M+H]+), a coumarin, was identified in SFR5, consistent with five fragment ions from the GNPS library. This coumarin is common in the plant kingdom, for example in Artemisia capillaris Herba (Asteraceae), which has shown interesting anticonvulsant activity in vivo [39]. However, it is the first description of esculetin (2) in Piper. Indeed, coumarins are not typically associated with the Piper genus. Nonetheless, some articles have suggested the presence of this class in Piper [40];
- (e)
- Three amides, 9-octadecenamide (76, m/z 282.2791, [M+H]+), 13-docosenamide (81, m/z 338.3416, [M+H]+), both with twelve fragment ions consistent with the GNPS library, and pipzorine (85, m/z 364.3574, [M+H]+), are present in SFR3, SFR4, and SFR5. The presence of pipzorine in SFR5 was inferred through manual annotation propagation, based on the spectra of the other two amides present in the same molecular cluster. Amides, including pipzorine, are commonly found in the Piper genus [20,41]. However, this is the first description of amides 76 and 81 for this genus;
- (f)
- The piperamides piperlonguminine (64, m/z 274.1453, [M+H]+) and piperine (67, m/z 286.1436, [M+H]+) were detected in SFR4 and SFR5, with eight and twelve fragment ions, respectively, consistent with the GNPS library. Piperlonguminine (64) has shown interesting in vivo antitumor activity [42], and piperine (67) exhibits various pharmacological effects, including antiproliferative, antitumor, antiangiogenic, antioxidant, antidiabetic, anti-obesity, cardioprotective, antimicrobial, anti-aging, and immunomodulatory properties in various in vitro and in vivo experimental assays [43]. Additionally, compound 67 has demonstrated antiparasitic, hepatoprotective, antiallergic, anti-inflammatory, and neuroprotective properties [44]. Piperamides are common in Piper [20,45]; however, this is the first description of these compounds in P. aduncum;
- (g)
- Other phenolic compounds such as vanillic acid (4, m/z 169.0498, [M+H]+), vanillin (6, m/z 153.0547, [M+H]+), sinapaldehyde (33, m/z 209.0808, [M+H]+), and ethyl vanillate (50, m/z 197.0811, [M+H]+) were detected in SFR4 and SFR5, all with six fragment ions corresponding to the GNPS library. Compounds 4 and 6 are widespread in the plant kingdom. Vanillic acid (4) is well-known for its pharmacological properties such as antioxidant, anti-inflammatory, immunostimulant, neuroprotective, hepatoprotective, cardioprotective, and antiapoptotic effects. It has also been reported to have the potential to attenuate Aβ1-42-induced cognitive impairment and oxidative stress, contributing to the treatment of Alzheimer’s disease [44]. Vanillin (6) also exhibits anticancer, antidiabetic, anti-inflammatory, and antimicrobial activities [46].
| Compound No. | Rt (min) | Precursor Ion (m/z) | Fragment Ions (MS2) * | Molecular Formula | Adduct Ion | Exact Mass (m/z) | Annotated Compound | Error (ppm) | Shared Peaks | Annotation Type |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.93 | 153.0551 | 153; 135; 121; 111; 109; 107; 94; 81 | C8H8O3 | [M+H]+ | 153.05516 | methyl 3-hydroxybenzoate | −0.4 | - | Manual inspection |
| 2 | 4.74 | 179.0343 | 179; 151; 147; 135; 133; 123 | C9H6O4 | [M+H]+ | 179.03443 | esculetin | −0.7 | 5 | GNPS library |
| 3 | 5.37 | 183.0664 | 183; 151; 139; 124; 107; 95; 79 | C9H10O4 | [M+H]+ | 183.06573 | ethyl vanillate | 3.7 | 5 | GNPS library |
| 4 | 5.40 | 169.0498 | 169; 151; 125; 111; 93; 65 | C8H8O4 | [M+H]+ | 169.05008 | vanillic acid | −1.7 | 6 | GNPS library |
| 5 | 6.80 | 213.0762 | 213; 181; 169; 154; 149; 137; 109; 91; 81 | C10H12O5 | [M+H]+ | 213.07629 | methyl 4-hydroxy-3,5-dimethoxybenzoate | −0.4 | 13 | GNPS library |
| 6 | 7.31 | 153.0547 | 153; 125; 111; 93; 65 | C8H8O3 | [M+H]+ | 153.05516 | vanillin | −3.0 | 6 | GNPS library |
| 7 | 7.65 | 197.0810 | 197; 179; 169; 165; 156; 147; 137; 119; 97; 95; 69 | C10H10O4 | [M+H]+ | 197.08138 | 1-propanone, 1-(3,5-dihydroxy-4-methoxyphenyl) | −1.9 | - | Manual inspection |
| 8 | 7.82 | 197.0814 | 197; 169; 165; 156; 147; 137; 119; 97; 69 | C10H12O4 | [M+H]+ | 197.08138 | 1-propanone, 1-(3,4-dihydroxy-5-methoxyphenyl) | 0.1 | - | Manual inspection |
| 9 | 8.71 | 207.1380 | 207; 189; 161; 149; 123; 95 | C13H18O2 | [M+H]+ | 207.1385 | not identified | −2.4 | - | - |
| 10 | 8.89 | 169.0496 | 169; 141; 137; 125; 111; 110; 107; 79 | C8H8O4 | [M+H]+ | 169.05008 | methyl 3,4-dihydroxybenzoate | −2.8 | 7 | GNPS library |
| 11 | 8.92 | 179.0703 | 179; 147; 119 | C10H12O4 | [M+H-H2O]+ | 179.07081 | dihydroferulic acid | −2.8 | - | Manual inspection |
| 12 | 8.93 | 389.2171 | 371; 227; 209; 191; 163; 149; 125; 107; 85; 69 | C19H32O8 | [M+H]+ | 389.21754 | 4-[3-(β-D-glucopyranosyloxy)butyl]-4-hydroxy-3,5,5-trimethyl-2-cyclohexen-1-one | −1.1 | 13 | GNPS library |
| 13 | 9.30 | 371.2065 | 371; 353; 209; 191; 125; 111 | C19H30O7 | [M+H]+ | 371.20697 | ranuncoside | −1.3 | - | Manual inspection |
| 14 | 9.44 | 179.0704 | 179; 147; 119 | C10H12O4 | [M+H-H2O]+ | 179.07081 | dihydroisoferulic acid | −2.3 | - | Manual inspection |
| 15 | 9.62 | 211.0964 | 211; 193; 179; 170; 147; 137; 123 | C11H14O4 | [M+H]+ | 211.09703 | 1-propanone, 1-(3,5-dimethoxy-4-hidroxyphenyl) | −3.0 | - | Manual inspection |
| 16 | 9.67 | 449.1069 | 449; 431; 413; 395; 383; 353, 339, 329; 299 | C21H20O11 | [M+H]+ | 449.10838 | orientin | −3.3 | 14 | GNPS library |
| 17 | 9.70 | 581.1502 | 581; 449; 431; 413; 383; 329; 299; | C26H28O15 | [M+H]+ | 581.15064 | 2-O-β-D-xylopyranosylisoorientin | −0.8 | 13 | GNPS library |
| 18 | 9.73 | 177.0546 | 177; 163; 149; 145; 135; 117; 89 | C10H10O4 | [M+H-H2O]+ | 177.05516 | ferulic acid | −3.2 | 7 | GNPS library |
| 19 | 9.77 | 195.0653 | 195; 177; 163; 145; 135; 117; 89 | C10H10O4 | [M+H]+ | 195.06573 | ferulic acid | −2.2 | 7 | GNPS library |
| 20 | 9.85 | 611.1610 | 611; 449; 431; 413; 395; 383; 353; 329; 311; 299; 287 | C27H30O16 | [M+H]+ | 611.1612 | 2-O-β-L-galactopyranosylorientin | −0.3 | 10 | GNPS library |
| 21 | 9.90 | 595.1657 | 595; 449; 413; 383; 353; 329; 299; 287 | C27H30O15 | [M+H]+ | 595.16629 | isoorientin 2″-O-rhamnoside | −1.0 | 7 | GNPS library |
| 22 | 9.90 | 246.1490 | 246; 217; 177; 164; 137; 83; 55 | C15H19NO2 | [M+H]+ | 246.1494 | not identified | −1.6 | - | - |
| 23 | 9.94 | 463.1236 | 463; 445; 427; 409; 397; 367; 353; 343; 313 | C22H22O11 | [M+H]+ | 463.12403 | swertiajaponin | −0.9 | 6 | GNPS library |
| 24 | 10.07 | 197.1174 | 197; 179; 161; 135; 133; 107; 93 | C11H16O3 | [M+H]+ | 197.11776 | loliolide | −1.8 | 7 | GNPS library |
| 25 | 10.09 | 565.1554 | 565; 433; 415; 397; 367; 337; 313; 283 | C26H28O14 | [M+H]+ | 565.15573 | 3′-hydroxypuerarin 2″-β-D-xyloside | −0.6 | 8 | GNPS library |
| 26 | 10.11 | 595.1658 | 595; 475; 433; 415; 397; 337; 313; 271; 85 | C27H30O15 | [M+H]+ | 595.16629 | isovitexin 2″-O-glucoside | −0.8 | 13 | GNPS library |
| 27 | 10.13 | 433.1127 | 433; 415; 397; 367; 349; 337; 313; 283 | C21H20O10 | [M+H]+ | 433.11347 | vitexin | −1.8 | 9 | GNPS library |
| 28 | 10.18 | 579.1700 | 579; 433; 415; 397; 337; 313; 271; 217; 85 | C27H30O14 | [M+H]+ | 579.17138 | vitexin 2″-O-rhamnoside | −2.4 | 13 | GNPS library |
| 29 | 10.32 | 433.1130 | 433; 415; 397; 367; 349; 337; 313; 283 | C21H20O10 | [M+H]+ | 433.11347 | isovitexin | −1.1 | 14 | GNPS library |
| 30 | 10.42 | 463.1232 | 463; 445; 427; 367; 343; 313; 261; 217; 151; 96 | C22H22O11 | [M+H]+ | 463.12403 | diosmetin 6-C-glucoside | −1.8 | - | Manual inspection |
| 31 | 10.43 | 593.1868 | 593; 447; 429; 381; 351; 327; 297; 285 | C28H32O14 | [M+H]+ | 593.18703 | acacetin 7-O-rutinoside | −0.4 | 4 | GNPS library |
| 32 | 10.53 | 447.1281 | 447; 429; 411; 393; 381; 351; 327; 297; 285 | C22H22O10 | [M+H]+ | 447.12912 | isoswertisin | −2.3 | - | Manual inspection |
| 33 | 10.60 | 209.0808 | 209; 194; 181; 177; 149; 145; 121; 55 | C11H12O4 | [M+H]+ | 209.08138 | sinapaldehyde | −2.8 | 6 | GNPS library |
| 34 | 10.65 | 387.2016 | 387; 355; 225; 207; 189; 167; 149; 123 | C19H30O8 | [M+H]+ | 387.20189 | roseoside | −0.7 | - | Manual inspection |
| 35 | 10.87 | 417.1184 | 417; 399; 381; 321; 297; 267;217; 167; 105 | C21H20O9 | [M+H]+ | 417.11855 | pueranin | −0.4 | 17 | GNPS library |
| 36 | 10.88 | 183.0665 | 183; 155; 151; 137; 124; 123; 111; 107; 93; 79 | C9H10O4 | [M+H]+ | 183.06573 | methyl 3-hydroxy-4-methoxybenzoate | 4.2 | 5 | GNPS library |
| 37 | 10.89 | 623.1972 | 623; 503; 461; 425; 365; 341; 299; 127; 85 | C29H34O15 | [M+H]+ | 623.19759 | embinoidin | −0.6 | - | Manual inspection |
| 38 | 10.94 | 433.1129 | 433; 415; 367; 337; 313; 283; 271 | C21H20O10 | [M+H]+ | 433.11347 | apigenin 7-O-glucoside | −1.3 | - | Manual inspection |
| 39 | 10.95 | 609.1814 | 609; 447; 429; 411; 381; 351; 327; 297; 285 | C28H32O15 | [M+H]+ | 609.18194 | swertisin-2″-O-glucoside | −0.9 | 9 | GNPS library |
| 40 | 10.96 | 593.1866 | 593; 447; 429; 381; 327; 297; 285; 85 | C28H32O14 | [M+H]+ | 593.18703 | swertisin-2″-O-rhamnoside | −0.7 | 4 | GNPS library |
| 41 | 11.07 | 447.1280 | 447; 429; 411; 381; 327; 297; 261; 162; 135; 96 | C22H22O10 | [M+H]+ | 447.12912 | 3′-methoxypuerarin | −2.5 | 13 | GNPS library |
| 42 | 11.20 | 461.1443 | 461; 341; 299 | C23H24O10 | [M+H]+ | 461.14477 | 7-(β-D-glucopyranosyloxy)-5-methoxy-2-(4-methoxyphenyl) 4H-1-benzopyran-4-one | −1.0 | - | Manual inspection |
| 43 | 11.38 | 211.1693 | 211; 193; 175; 151; 135; 109; 95; 69 | C13H22O2 | [M+H]+ | 211.1698 | not identified | −2.4 | - | - |
| 44 | 11.52 | 195.1016 | 195; 167; 163; 154; 135; 107; 103; 91; 79 | C11H14O3 | [M+H]+ | 195.10211 | 6-methoxy eugenol | −2.6 | 6 | GNPS library |
| 45 | 11.68 | 447.1287 | 447; 429; 411; 393; 381; 351; 327; 297; 285 | C22H22O10 | [M+H]+ | 447.12912 | swertisin | −0.9 | 11 | GNPS library |
| 46 | 11.68 | 207.0652 | 207; 192; 179; 175; 147; 119; 91 | C11H12O5 | [M+H-H2O]+ | 207.06573 | trans-sinapic acid | −2.6 | 11 | GNPS library |
| 47 | 11.78 | 461.1466 | 461; 443; 425; 407; 395; 365; 351; 341; 311; 159; 109 | C23H24O10 | [M+H]+ | 461.14477 | embigenin | 4.0 | - | Manual inspection |
| 48 | 11.81 | 179.0703 | 179; 151; 147; 137; 123; 119; 105; 91 | C10H10O3 | [M+H]+ | 179.07081 | coniferaldehyde | −2.8 | 14 | GNPS library |
| 49 | 11.82 | 177.0545 | 177; 163; 149; 145; 135; 117; 89 | C10H10O4 | [M+H-H2O]+ | 177.05516 | isoferulic acid | −3.7 | 6 | GNPS library |
| 50 | 11.86 | 197.0811 | 197; 169; 151; 125; 111; 93; 65 | C10H12O4 | [M+H]+ | 197.08138 | ethyl vanillate | −1.4 | 7 | GNPS library |
| 51 | 11.89 | 489.1389 | 489; 471; 453; 411; 393; 327; 297; 121; 96 | C24H24O11 | [M+H]+ | 489.13968 | 2′-O-acetyl-7-O-methyl vitexin | −1.6 | - | Manual inspection |
| 52 | 12.19 | 207.0653 | 207; 192; 179; 175; 147; 119; 91 | C11H12O5 | [M+H-H2O]+ | 207.06573 | cis-sinapic acid | −2.1 | 10 | GNPS library |
| 53 | 12.33 | 447.1291 | 447; 429; 411; 393; 381; 351; 327; 297; 285 | C22H22O10 | [M+H]+ | 447.12912 | 6-β-D-glucopyranosyl-7-hydroxy-2-(4-hydroxyphenyl)-5-methoxy-4H-1-benzopyran-4-one | 0.0 | 10 | GNPS library |
| 54 | 12.45 | 271.0959 | 271; 229; 167; 131; 103 | C16H14O4 | [M+H]+ | 271.09703 | alpinetin | −4.2 | 5 | GNPS library |
| 55 | 12.56 | 221.1172 | 221; 189; 165; 153; 109; 69 | C13H16O3 | [M+H]+ | 221.11776 | 4-methoxy-3-(3′-methyl-2′-butenyl)-benzoic acid | −2.5 | - | Manual inspection |
| 56 | 12.56 | 207.1012 | 207; 165; 151; 107; 69 | C12H14O3 | [M+H]+ | 207.10211 | 4-hydroxy-3-(3′-methyl-2′-butenyl)-benzoic acid | −4.4 | - | Manual inspection |
| 57 | 12.59 | 273.1124 | 273; 255; 245; 217; 169; 141; 133; 105; 91 | C16H16O4 | [M+H]+ | 273.11268 | 2,6-dihydroxy-4-methoxydihydrochalcone | −1.0 | 8 | GNPS library |
| 58 | 12.61 | 237.1848 | 237; 219; 201; 191; 159; 145; 135; 121; 95; 81 | C15H24O2 | [M+H]+ | 237.18545 | bisabolene-1,4-endoperoxide | −2.7 | 10 | GNPS library |
| 59 | 12.71 | 275.2006 | 275; 257; 239; 161; 147; 133; 119; 105; 91; | C18H28O3 | [M+H-H2O]+ | 275.2011 | (10E,12Z,15Z)-9-oxooctadeca-10,12,15-trienoic acid | −1.8 | 8 | GNPS library |
| 60 | 12.73 | 293.2109 | 293; 275; 257; 239; 189; 133; 107; 95; 81; 67 | C18H28O3 | [M+H]+ | 293.21166 | not identified | −2.6 | - | - |
| 61 | 12.75 | 311.2216 | 311; 293; 275; 257; 189; 121; 109; 95; 81; 67 | C18H32O5 | [M+H-H2O]+ | 311.22223 | 9,12,13-trihydroxyoctadeca-10,15-dienoic acid | −2.0 | 9 | GNPS library |
| 62 | 12.89 | 317.1020 | 317; 193; 185; 177; 167; 145; | C17H16O6 | [M+H]+ | 317.10251 | Eriodictyol-7,3′-dimethyl ether | −1.6 | 7 | GNPS library |
| 63 | 12.90 | 287.0909 | 287; 269; 167; 147; 119; | C16H14O5 | [M+H]+ | 287.09194 | sakuranetin | −3.6 | 7 | GNPS library |
| 64 | 12.97 | 274.1453 | 274; 201; 171; 159; 143; 135; 115 | C16H19NO3 | [M+H]+ | 274.14431 | piperlonguminine | 3.6 | 8 | GNPS library |
| 65 | 12.98 | 257.0808 | 257; 212; 171; 153; 131 | C15H12O4 | [M+H]+ | 257.08138 | pinocembrine | −2.3 | 6 | GNPS library |
| 66 | 13.17 | 285.1134 | 285; 243; 181; 131; 91 | C17H16O4 | [M+H]+ | 285.11268 | 5,7-dimethoxyflavanone | 2.5 | 5 | GNPS library |
| 67 | 13.22 | 286.1436 | 286; 201; 171; 143; 135; 112; 84 | C17H19NO3 | [M+H]+ | 286.14431 | piperine | −2.5 | 12 | GNPS library |
| 68 | 13.25 | 273.1497 | 273; 217; 199; 173; 159; 91; 69 | C17H20O3 | [M+H]+ | 273.14906 | 2,2-dimethyl-8-(3-methylbut-2-en-1-yl)-2H-chromene-6-carboxylic acid | 2.3 | - | Manual inspection |
| 69 | 13.27 | 285.0758 | 285; 270; 242; 222; 139; 99; 68 | C16H12O5 | [M+H]+ | 285.07629 | wogonin | −1.7 | - | Manual inspection |
| 70 | 13.48 | 287.1640 | 287; 231; 219; 175; 157; 105; 69 | C18H22O3 | [M+H]+ | 287.16471 | 4-methoxy-3-(3-methylbut-2-en-1-yl)-5-(3-methylbuta-1,3-dien-1-yl)benzoic acid | −2.5 | - | Manual inspection |
| 71 | 13.50 | 275.2005 | 275; 239; 161; 147; 133; 119;105 | C18H28O3 | [M+H-H2O]+ | 275.2011 | 9-oxo-10,12,15-octadecatrienoic acid | −2.2 | 14 | GNPS library |
| 72 | 13.52 | 273.1118 | 273; 255; 223; 177; 133; 115; 105; 91; | C16H16O4 | [M+H]+ | 273.11268 | 2,4-dihydroxy-6-methoxydihydrochalcone | −3.2 | 8 | GNPS library |
| 73 | 13.53 | 289.1797 | 289; 257; 233; 221; 165; 153; 69 | C18H24O3 | [M+H]+ | 289.18036 | (3′,7′-dimethyl-2′,6′-octadienyl)-4-methoxybenzoic acid | −2.3 | - | Manual inspection |
| 74 | 13.55 | 235.1328 | 235; 207; 189; 151; 107; 69 | C14H18O3 | [M+H]+ | 235.13341 | methyl 4-metoxy-3-(3′-methyl-2′-butenyl)benzoate | −2.6 | - | Manual inspection |
| 75 | 13.76 | 351.2528 | 351; 259; 241; 161; 147; 133; 93; 81; 67 | C21H34O4 | [M+H]+ | 351.25353 | 2,3-dihydroxypropyl-6,9,12,15-octadecatetraenoate | −2.1 | - | Manual inspection |
| 76 | 13.81 | 282.2791 | 282; 265;247; 163; 149; 135; 97; 83; 69; 55 | C18H35NO | [M+H]+ | 282.27968 | 9-octadecenamide | −2.1 | 12 | GNPS library |
| 77 | 13.96 | 277.2160 | 277; 221; 163; 149; 135; 121; 107; 93; 79 | C18H30O3 | [M+H-H2O]+ | 277.21675 | 9-hydroxy-10,12,15-octadecatrienoic acid | −2.7 | 9 | GNPS library |
| 78 | 14.16 | 279.2318 | 279; 209; 173; 137; 123; 109; 95; 81; 67 | C18H32O3 | [M+H-H2O]+ | 279.2324 | 9,10-epoxyoctadecenoic acid | −2.1 | 7 | GNPS library |
| 79 | 14.30 | 427.3890 | 427; 324; 199; 71; 67; | C25H50N2O3 | [M+H]+ | 427.38996 | isostearamidopropyl betaine ** | −2.2 | - | Manual inspection |
| 80 | 14.32 | 357.3000 | 357; 339; 283; 265; 247; 149; 135; 121; 95; 81; 69 | C21H40O4 | [M+H]+ | 357.30048 | monoolein | −1.3 | 12 | GNPS library |
| 81 | 14.41 | 338.3416 | 338; 321; 303; 135; 97; 83; 69; 55 | C22H43NO | [M+H]+ | 338.34229 | 13-docosenamide | −2.0 | 12 | GNPS library |
| 82 | 14.55 | 305.2472 | 305; 163; 149; 135; 121; 107; 93; 79; 67; 55 | C20H34O3 | [M+H-H2O]+ | 305.24805 | 15-oxo-11(Z),13(E)-eicosadienoic acid | −2.8 | 9 | GNPS library |
| 83 | 14.73 | 372.3469 | 372; 354; 311; 106; 88; 70 | C22H45NO3 | [M+H]+ | 372.34776 | stearic diethanolamide ** | −2.3 | - | Manual inspection |
| 84 | 14.74 | 293.2474 | 293; 261; 243; 137; 123; 109; 95; 81; 67 | C19H34O3 | [M+H-H2O]+ | 293.24805 | 13-hydroxy-9(Z),11(E)-octadecadienoic acid, methyl ester | −2.2 | 9 | GNPS library |
| 85 | 14.76 | 364.3574 | 364; 282; 247; 121; 97; 83; 69; 55 | C24H45NO | [M+H]+ | 364.35794 | pipzorine | −1.5 | - | Manual inspection |
| 86 | 14.82 | 353.2683 | 353; 261; 243; 233; 173; 121; 109; 95; 81 | C21H36O4 | [M+H]+ | 353.26918 | monolinolenin | −2.5 | 10 | GNPS library |
| 87 | 14.94 | 470.4203 | 470; 288; 270; 227; 106; 88 | C28H55NO4 | [M+H]+ | 470.42093 | not identified | −1.3 | - | - |
| 88 | 15.31 | 355.2840 | 355; 337; 263; 245; 161; 121; 109; 95; 81; 67 | C21H38O4 | [M+H]+ | 355.28483 | monolinolein | −2.3 | 8 | GNPS library |
| 89 | 15.52 | 307.2627 | 307; 261; 243; 137; 123; 109; 95; 81; 67 | C20H34O2 | [M+H]+ | 307.2637 | 9(Z),11(E),13(E)-octadecatrienoic acid, ethyl ester | −3.3 | 6 | GNPS library |
3. Materials and Methods
3.1. Plant Material
3.2. Extract Preparation and Column Chromatography of the Bioactive Partition
3.3. Analysis by Ultra-High-Performance Liquid Chromatography Coupled with High-Resolution Mass Spectrometry in Tandem (UHPLC-HRMS/MS)
3.4. Processing of UHPLC-HRMS/MS Data by Molecular Network
3.5. Biological Assay
3.5.1. Mycobacterium tuberculosis H37Rv Growth
3.5.2. Growth Inhibition Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Posada, R.C. Piperaceae. In Flora Mesoamericana; Davidse, G., Ulloa, C., Hernández, H., Eds.; Universidad Nacional Autónoma de México : Mexico City, Mexico; Missouri Botanical Garden Press: St. Louis, MO, USA; Natural History Museum: London, UK, 2020; Volume 2, p. 618. [Google Scholar]
- Piper in Flora and Funga of Brasil. Rio de Janeiro Botanical Garden. Available online: https://floradobrasil.jbrj.gov.br/FB12738 (accessed on 15 January 2024).
- Queiroz, G.A.; Guimarães, E.F. Piper L. (Piperaceae) from Eastern Metropolitan, RJ, Brazil. Braz. J. Dev. 2020, 6, 93597–93634. [Google Scholar] [CrossRef]
- Monzote, L.; Scull, R.; Cos, P.; Setzer, W.N. Essential Oil from Piper aduncum: Chemical Analysis, Antimicrobial Assessment, and Literature Review. Medicines 2017, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Zakaria, Z.; Gyawali, R.; Ibrahim, S.; Rajkovic, J.; Shinwari, Z.; Khan, T.; Sharifi-Rad, J.; Ozleyen, A.; Turkdonmez, E.; et al. Piper Species: A Comprehensive Review on Their Phytochemistry, Biological Activities and Applications. Molecules 2019, 24, 1364. [Google Scholar] [CrossRef] [PubMed]
- Durofil, A.; Radice, M.; Blanco-Salas, J.; Ruiz-Téllez, T. Piper aduncum essential oil: A promising insecticide, acaricide and antiparasitic. A review. Parasite 2021, 28, 42. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Jensen, H.; Philogène, B.; Arnason, J. A review of Piper spp. (Piperaceae) phytochemistry, insecticidal activity and mode of action. Phytochem. Rev. 2007, 7, 65–75. [Google Scholar]
- Silva, J.; Trindade, R.; Alves, N.; Figueiredo, P.; Maia, J.; Setzer, W. Essential Oils from Neotropical Piper Species and Their Biological Activities. Int. J. Mol. Sci. 2017, 18, 2571. [Google Scholar] [CrossRef]
- Peixoto, J.F.; Ramos, Y.J.; Moreira, D.L.; Alves, C.R.; Gonçalves-Oliveira, L.F. Potential of Piper spp. as a source of new compounds for the leishmaniases treatment. Parasit. Res. 2021, 1, 2731–2747. [Google Scholar] [CrossRef]
- Moreira, D.L.; Guimarães, E.F.; Kaplan, M.A.C. A chromene from Piper aduncum. Phytochemistry 1998, 48, 1075–1077. [Google Scholar] [CrossRef]
- Lock, O.; Rojas, R. Chemistry and Pharmacology of Piper aduncum L. (“Matico”). Rev. Quím. 2004, 8, 27–32. [Google Scholar]
- Pohlit, A.M.; Pinto, A.C.S.; Mause, R. Piper aduncum L.: Pluripotent Plant and Source of Important Phytochemical Substances. Fitos Mag. 2006, 2, 7–18. [Google Scholar] [CrossRef]
- Lago, J.H.G.; Chen, A.; Young, M.C.M.; Guimaraes, E.F.; Oliveira, A.; Kato, M.J. Prenylated benzoic acid derivatives from Piper aduncum L. and P. hostmannianum C. DC. (Piperaceae). Phytochem. Lett. 2009, 2, 96–98. [Google Scholar] [CrossRef]
- Pacheco, F.V.; Alvarenga, I.C.A.; Junior, P.M.R.; Pereira Pinto, J.E.B.; de Paula Avelar, R.; Alvarenga, A.A. Growth and production of secondary compounds in monkey-pepper (’Piper aduncum’ L.) leaves cultivated under altered ambient light. Aust. J. Crop Sci. 2014, 8, 1510–1516. [Google Scholar]
- Fazolin, M.; Monteiro, A.F.M.; Bizzo, H.R.; Gama, P.E.; Viana, L.O.; Lima, M.É.C. Insecticidal activity of Piper aduncum oil: Variation in dillapiole content and chemical and toxicological stability during storage. Acta Amazon. 2021, 52, 179–188. [Google Scholar] [CrossRef]
- Assunção, J.A.; Silva, M.D.; Felisberto, J.R.S.; Guimarães, E.F.; de Azevedo, Q.G.; Ramos, Y.J.; Moreira, D.L. Effects of postharvest UV-C irradiation on essential oils from leaves of Piper aduncum L. for industrial and medicinal use. Ind. Crop. Prod. 2023, 203, 117216. [Google Scholar] [CrossRef]
- WHO. Global Tuberculosis Report; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Demarque, D.P.; Crotti, A.E.; Vessecchi, R.; Lopes, J.L.; Lopes, N.P. Fragmentation reactions using electrospray ionization mass spectrometry: An important tool for the structural elucidation and characterization of synthetic and natural products. Nat. Prod. Rep. 2016, 33, 432–455. [Google Scholar] [CrossRef] [PubMed]
- Parmar, V.S.; Jain, S.C.; Bisht, K.S.; Jain, R.; Taneja, P.; Jha, A.; Tyagi, D.; Prasad, A.K.; Wengel, J.; Olsen, C.E.; et al. Phytochemistry of the genus Piper. Phytochemistry 1997, 46, 597–673. [Google Scholar] [CrossRef]
- Masuoka, C.; Ono, M.; Ito, Y.; Nohara, T. Antioxidative, Antihyaluronidase and Antityrosinase Activities of Some Constituents from the Aerial Part of Piper elongatum VAHL. Food Sci. Technol. Res. 2009, 9, 197–201. [Google Scholar] [CrossRef]
- Moreira, D.L.; Guimarães, E.F.; Kaplan, M.C. A C-glucosylflavone from leaves of Piper lhotzkyanum. Phytochemistry 2000, 55, 783–786. [Google Scholar] [CrossRef]
- Thao, N.P.; Luyen, B.T.T.; Widowati, W.; Fauziah, N.; Maesaroh, M.; Herlina, T.; Manzoor, Z.; Ali, I.; Koh, Y.S.; Kim, Y.H. Anti-inflammatory flavonoid C-glycosides from Piper aduncum leaves. Planta Med. 2016, 82, 1475–1481. [Google Scholar] [CrossRef]
- de Rijke, E.; Out, P.; Niessen, W.M.A.; Ariese, F.; Gooijer, C.; Brinkman, U.A.T.H. Analytical separation and detection methods for flavonoids. J. Chromatogr. A 2006, 1112, 31–63. [Google Scholar] [CrossRef] [PubMed]
- Ticona, J.C.; Bilbao-Ramos, P.; Amesty, Á.; Flores, N.; Dea-Ayuela, M.A.; Bazzocchi, I.L.; Jiménez, I.A. Flavonoids from Piper Species as Promising Antiprotozoal Agents against Giardia intestinalis: Structure-Activity Relationship and Drug-Likeness Studies. Pharmaceuticals 2022, 15, 1386. [Google Scholar] [CrossRef] [PubMed]
- Minh, T.N.; Van, T.M.; Khanh, T.D.; Xuan, T.D. Isolation and identification of constituents exhibiting antioxidant, antibacterial, and antihyperuricemia activities in Piper methysticum root. Foods 2022, 11, 3889. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Tian, J.; Li, Y.; Liu, M.; Chao, Y.; Cai, Y.; Zheng, G.; Fang, Y. Identification of components in Citri Sarcodactylis Fructus from different origins via UPLC-Q-Exactive Orbitrap/MS. ACS Omega 2021, 6, 17045–17057. [Google Scholar] [CrossRef] [PubMed]
- Sruthi, D.; Zachariah, T.J. Phenolic profiling of Piper species by liquid chromatography-mass spectrometry. J. Spices Aromat. Crop. 2016, 25, 123–132. [Google Scholar]
- Fan, R.; Tao, X.Y.; Xia, Z.Q.; Sim, S.; Hu, L.S.; Wu, B.D.; Wang, Q.H.; Hao, C.Y. Comparative transcriptome and metabolome analysis of resistant and susceptible Piper species upon infection by the oomycete Phytophthora capsici. Front. Plant Sci. 2022, 13, 864927. [Google Scholar] [CrossRef]
- Baldoqui, D.C.; Kato, M.J.; Cavalheiro, A.J.; Bolzani, V.S.; Young, M.C.M.; Furlan, M. A chromene and prenylated benzoic acid from Piper aduncum. Phytochemistry 1999, 51, 899–902. [Google Scholar] [CrossRef]
- Rossa, G.E.; Almeida, R.N.; Vargas, R.M.F.; Cassel, E.; Moyna, G. Sequential extraction methods applied to Piper hispidinervum: An improvement in the processing of natural products. Can. J. Chem. Eng. 2018, 96, 756–762. [Google Scholar] [CrossRef]
- Su, H.; Hu, Y.J.; Mo, X.Y.; Diao, N.; Wang, Y.; Liang, D. Three new phenylpropanoid glucosides and a new tyramine-type alkamide from Piper puberulum. J. Asian Nat. Prod. Res. 2023, 1, 11. [Google Scholar] [CrossRef]
- Quílez, A.; Berenguer, B.; Gilardoni, G.; Souccar, C.; De Mendonça, S.; Oliveira, L.F.S.; Martín-Calero, M.J.; Vidari, G. Anti-secretory, anti-inflammatory and anti-Helicobacter pylori activities of several fractions isolated from Piper carpunya Ruiz & Pav. J. Ethnopharmacol. 2010, 128, 583–589. [Google Scholar]
- Flanigan, P.M., IV; Brady, J.J.; Judge, E.J.; Levis, R.J. Determination of inorganic improvised explosive device signatures using laser electrospray mass spectrometry detection with offline classification. Anal. Chem. 2011, 83, 7115–7122. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Yan, G.-Y.; Wu, J.-L.; Zong, X.; Liu, Z.; Zhou, H.; Liu, L.; Li, N. Qualitative and quantitative analysis of lipo-alkaloids and fatty acids in Aconitum carmichaelii using LC-MS and GC-MS. Phytochem. Anal. 2018, 29, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Oda, Y.; Hirota, M. (10E, 12Z, 15Z)-9-hydroxy-10, 12, 15-octadecatrienoic acid methyl ester as an anti-inflammatory compound from Ehretia dicksonii. Biosci. Biotechnol. Biochem. 2000, 64, 882–886. [Google Scholar] [CrossRef]
- Liu, T.; Liang, Q.; Zhang, X.M.; Li, G.Q.; Xu, B.; Xu, W.H. Chemical constituents from Piper boehmeriifolium (Miq.) Wall. ex C. DC. Biochem. Syst. Ecol. 2017, 75, 27–30. [Google Scholar] [CrossRef]
- Torres-Santos, E.C.; Moreira, D.L.; Kaplan, M.A.C.; Meirelles, M.N.; Rossi-Bergmann, B. Selective effect of 2′, 6′-dihydroxy-4′-methoxychalcone isolated from Piper aduncum on Leishmania amazonensis. Antimicrob. Agents Chemother. 1999, 43, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Torres-Santos, E.C.; Rodrigues, J.M., Jr.; Moreira, D.L.; Kaplan, M.A.C.; Rossi-Bergmann, B. Improvement of in vitro and in vivo antileishmanial activities of 2′, 6′-dihydroxy-4′-methoxychalcone by entrapment in poly (D, L-lactide) nanoparticles. Antimicrob. Agents Chemother. 1999, 43, 1776–1778. [Google Scholar] [PubMed]
- Woo, T.S.; Yoon, S.Y.; dela Penã, I.C.; Choi, Y.J.; Lee, H.L.; Choi, Y.J.; Lee, Y.S.; Ryu, J.H.; Choi, J.S.; Cheong, J.H. Anticonvulsant effect of Artemisia capillaris Herba in mice. Biomol. Ther. 2011, 19, 342–347. [Google Scholar] [CrossRef]
- Martins, S.A.; dos Santos, R.C.; Ramos, A.R.; Figueiredo, P.L.B.; da Silva, C.R.C.; da Silva, J.K.R. Allelopathic potential and phytochemical screening of Piper divaricatum extracts on germination and growth of indicator plant (Lactuca sativa). S. Afr. J. Bot. 2021, 138, 495–499. [Google Scholar] [CrossRef]
- Marques, A.M.; Kaplan, M.A.C. Active metabolites of the genus Piper against Aedes aegypti: Natural alternative sources for dengue vector control. Univ. Sci. 2015, 20, 61–82. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Pessoa, C.; Moraes, M.O.D.; Alencar, N.M.D.; Mesquita, R.O.; Lima, M.W.; Alves, A.P.N.N.; Pessoa, O.D.L.; Chaves, J.H.; Silveira, E.R.; et al. In vivo growth inhibition of sarcoma 180 by piperlonguminine, an alkaloid amide from the Piper species. J. Appl. Toxicol. 2008, 28, 599–607. [Google Scholar] [CrossRef]
- Kumar, S.; Kamboj, J.; Sharma, S. Overview for various aspects of the health benefits of Piper longum linn. fruit. J. Acupunct. Meridian Stud. 2011, 4, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Imran, M.; Nadeem, M.; Tufail, T.; Gondal, T.A.; Mubarak, M.S. Piperine: A review of its biological effects. Phytother. Res. 2021, 35, 680–700. [Google Scholar] [CrossRef]
- Sharma, N.; Tiwari, N.; Vyas, M.; Khurana, N.; Muthuraman, A.; Utreja, P. An overview of therapeutic effects of vanillic acid. Plant Arch. 2020, 20, 3053–3059. [Google Scholar]
- Olatunde, A.; Mohammed, A.; Ibrahim, M.A.; Tajuddeen, N.; Shuaibu, M.N. Vanillin: A food additive with multiple biological activities. Eur. J. Med. Chem. Rep. 2022, 5, 100055. [Google Scholar] [CrossRef]
- Mazlun, M.H.; Sabran, S.F.; Mohamed, M.; Bakar, M.F.A.; Abdullah, Z. Phenolic compounds as promising drug candidates in tuberculosis therapy. Molecules 2019, 24, 2449. [Google Scholar] [CrossRef]
- Askun, T. Investigation of Anti-Mycobacterial Activity of Orientin and Vitexin on the Six Mycobacterium tuberculosis Strains. Eur. J. Biol. 2023, 82, 124–131. [Google Scholar]
- Solnier, J.; Martin, L.; Bhakta, S.; Bucar, F. Flavonoids as novel efflux pump inhibitors and antimicrobials against both environmental and pathogenic intracellular mycobacterial species. Molecules 2020, 25, 734. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiang, X.; Gao, F.; Song, J.; Sun, J.; Wang, L.; Sun, X.; Lu, Z.; Zhang, H. Identification of plant-derived natural products as potential inhibitors of the Mycobacterium tuberculosis proteasome. BMC Complement. Med. Ther. 2014, 14, 400. [Google Scholar] [CrossRef]
- Costa-Oliveira, C.; Ramos, Y.J.; Queiroz, G.A.; Guimarães, E.F.; Heggdorne-Araújo, M.; Lassounskaia, E.B.; Muzitano, M.F.; Marcelino, D.S.; Moreira, D.L. Antimycobacterial Activity and Chemical Characterization of the Essential Oils from Reproductive Organs of Piper lhotzkyanum Kunth (Piperaceae). Rev. Virtual Quim. 2021, 5, 1196–1202. [Google Scholar] [CrossRef]
- Bernuci, K.Z.; Iwanaga, C.C.; Fernandez-Andrade, C.M.M.; Lorenzetti, F.B.; Torres-Santos, E.C.; Faiões, V.D.S.; Gonçalves, J.E.; Do Amaral, W.; Deschamps, C.; Scodro, R.B.L.; et al. Evaluation of chemical composition and antileishmanial and antituberculosis activities of essential oils of Piper species. Molécules 2016, 21, 1698. [Google Scholar] [CrossRef]
- Ramos, Y.J.; Costa-Oliveira, C.; Fonseca, I.C.; Marcelino, D.S.; Heggdorne-Araujo, M.; Lassounskaia, E.B.; Muzitano, M.F.; Queiroz, G.A.; Guimarães, E.F.; Moreira, D.L. Piper multinodum C.DC. (Piperaceae) essential oils chemical variation and biological activity against Mycobacterium tuberculosis. J. Med. Plant Res. 2021, 15, 413–422. [Google Scholar]
- Tekwu, E.M.; Askun, T.; Kuete, V.; Nkengfack, A.E.; Nyasse, B.; Etoa, F.X.; Beng, V.P. Antibacterial activity of selected Cameroonian dietary spices ethno-medically used against strains of Mycobacterium tuberculosis. J. Ethnopharmacol. 2012, 142, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Hussain, K.; Ismail, Z.; Sadikun, A.; Ibrahim, P. Antioxidant, anti-TB activities, phenolic and amide contents of standardised extracts of Piper sarmentosum Roxb. Nat. Prod. Res. 2009, 23, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Rukachaisirikul, T.; Siriwattanakit, P.; Sukcharoenphol, K.; Wongvein, C.; Ruttanaweang, P.; Wongwattanavuch, P.; Suksamrarn, A. Chemical constituents and bioactivity of Piper sarmentosum. J. Ethnopharmacol. 2004, 93, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Rukachaisirikul, T.; Prabpai, S.; Kongsaeree, P.; Suksamrarn, A. (+)-Bornyl piperate, a new monoterpene ester from Piper aff. pedicellatum roots. Chem. Pharm. Bull. 2004, 52, 760–761. [Google Scholar] [CrossRef] [PubMed]
- Rukachaisirikul, T.; Prabpai, S.; Champung, P.; Suksamrarn, A. Chabamide, a novel piperine dimer from stems of Piper chaba. Planta Med. 2002, 68, 853–855. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cheng, M.J.; Wu, C.C.; Peng, C.F.; Huang, H.Y.; Chang, H.S.; Wang, C.J.; Chen, I.S. Three new phenylpropanoids from the roots of Piper taiwanense and their inhibitory activities on platelet aggregation and Mycobacterium tuberculosis. Chem. Biodivers. 2014, 11, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Mata, R.; Morales, I.; Pérez, O.; Rivero-Cruz, I.; Acevedo, L.; Enriquez-Mendoza, I.; Tchau, R.; Franzblau, S.; Timmermann, B. Antimycobacterial Compounds from Piper sanctum. J. Nat. Prod. 2004, 67, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Scodro, R.B.L.; Pires, C.T.A.; Carrara, V.S.; Lemos, C.O.T.; Cardozo-Filho, L.; Souza, V.A.; Corrêa, A.G.; Siqueira, V.L.D.; Lonardoni, M.V.C.; Cardoso, R.F.; et al. Anti-tuberculosis neolignans from Piper regnellii. Phytomedicine 2013, 20, 600–604. [Google Scholar] [CrossRef]
- Scodro, R.B.D.L.; Espelho, S.C.; Pires, C.T.A.; Garcia, V.A.D.S.; Cardozo-Filho, L.; Cortez, L.E.R.; Pilau, E.J.; Ferracioli, K.R.C.; Siquiera, V.L.D.; Cardoso, R.F.; et al. A new benzoic acid derivative from Piper diospyrifolium and its anti-Mycobacterium tuberculosis activity. Phytochem. Lett. 2015, 11, 18–23. [Google Scholar] [CrossRef]
- Fernandez, C.M.M.; Baldin, V.P.; Ieque, A.L.; Bernuci, K.Z.; Almeida, R.T.; Valone, L.M.; Fonseca, D.P.; Makimori, R.Y.; Andrade, J.P.P.; Pilau, E.J.; et al. Anti-Mycobacterium tuberculosis activity of dichloromethane extract of Piper corcovadensis (Miq.) C. DC. roots and isolated compounds. Ind. Crop. Prod. 2015, 131, 341–347. [Google Scholar] [CrossRef]
- Hegeto, L.A.; Caleffi-Ferracioli, K.R.; Nakamura-Vasconcelos, S.S.; de Almeida, A.L.; Baldin, V.P.; Nakamura, C.V.; Siqueira, V.L.D.; Scodro, R.B.L.; Cardoso, R.F. In vitro combinatory activity of piperine and anti-tuberculosis drugs in Mycobacterium tuberculosis. Tuberculosis 2018, 111, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Flores, R.; Gupta, S.; Tamez-Guerra, R.; Mehta, R.T. Determination of MIC’s for Mycobacterium avium–M. intracellulare complex in liquid medium by a colorimetric method. J. Clin. Microbiol. 1995, 33, 1842–1846. [Google Scholar]



| Sample | MIC50 (µg/mL) |
|---|---|
| PAEEL | 121.6 ± 1.04 |
| PAHPL | 60.65 ± 1.05 |
| PADPL | 39.74 ± 1.02 |
| PAEPL | 27.98 ± 1.01 |
| PABPL | >128 |
| PAEES | >128 |
| PAHPS | 29.74 ± 1.03 |
| PADPS | 59.05 ± 1.04 |
| PAEPS | 145.40 ± 1.05 |
| PABPS | >128 |
| PAEEI | >128 |
| PAHPI | 48.61 ± 1.06 |
| PADPI | >128 |
| PAEPI | >128 NC |
| PABPI | >128 NC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Luna, A.V.; Fagundes, T.d.S.F.; Ramos, Y.J.; de Araújo, M.H.; Muzitano, M.F.; Calixto, S.D.; Simão, T.L.B.V.; de Queiroz, G.A.; Guimarães, E.F.; Marques, A.M.; et al. UHPLC-HRMS/MS Chemical Fingerprinting of the Bioactive Partition from Cultivated Piper aduncum L. Molecules 2024, 29, 1690. https://doi.org/10.3390/molecules29081690
de Luna AV, Fagundes TdSF, Ramos YJ, de Araújo MH, Muzitano MF, Calixto SD, Simão TLBV, de Queiroz GA, Guimarães EF, Marques AM, et al. UHPLC-HRMS/MS Chemical Fingerprinting of the Bioactive Partition from Cultivated Piper aduncum L. Molecules. 2024; 29(8):1690. https://doi.org/10.3390/molecules29081690
Chicago/Turabian Stylede Luna, Adélia Viviane, Thayssa da Silva Ferreira Fagundes, Ygor Jessé Ramos, Marlon Heggdorne de Araújo, Michelle Frazão Muzitano, Sanderson Dias Calixto, Thatiana Lopes Biá Ventura Simão, George Azevedo de Queiroz, Elsie Franklin Guimarães, André Mesquita Marques, and et al. 2024. "UHPLC-HRMS/MS Chemical Fingerprinting of the Bioactive Partition from Cultivated Piper aduncum L." Molecules 29, no. 8: 1690. https://doi.org/10.3390/molecules29081690
APA Stylede Luna, A. V., Fagundes, T. d. S. F., Ramos, Y. J., de Araújo, M. H., Muzitano, M. F., Calixto, S. D., Simão, T. L. B. V., de Queiroz, G. A., Guimarães, E. F., Marques, A. M., & Moreira, D. d. L. (2024). UHPLC-HRMS/MS Chemical Fingerprinting of the Bioactive Partition from Cultivated Piper aduncum L. Molecules, 29(8), 1690. https://doi.org/10.3390/molecules29081690