The DBB Family in Populus trichocarpa: Identification, Characterization, Evolution and Expression Profiles
Abstract
1. Introduction
2. Results
2.1. Identification of PtrDBB Genes in Populus
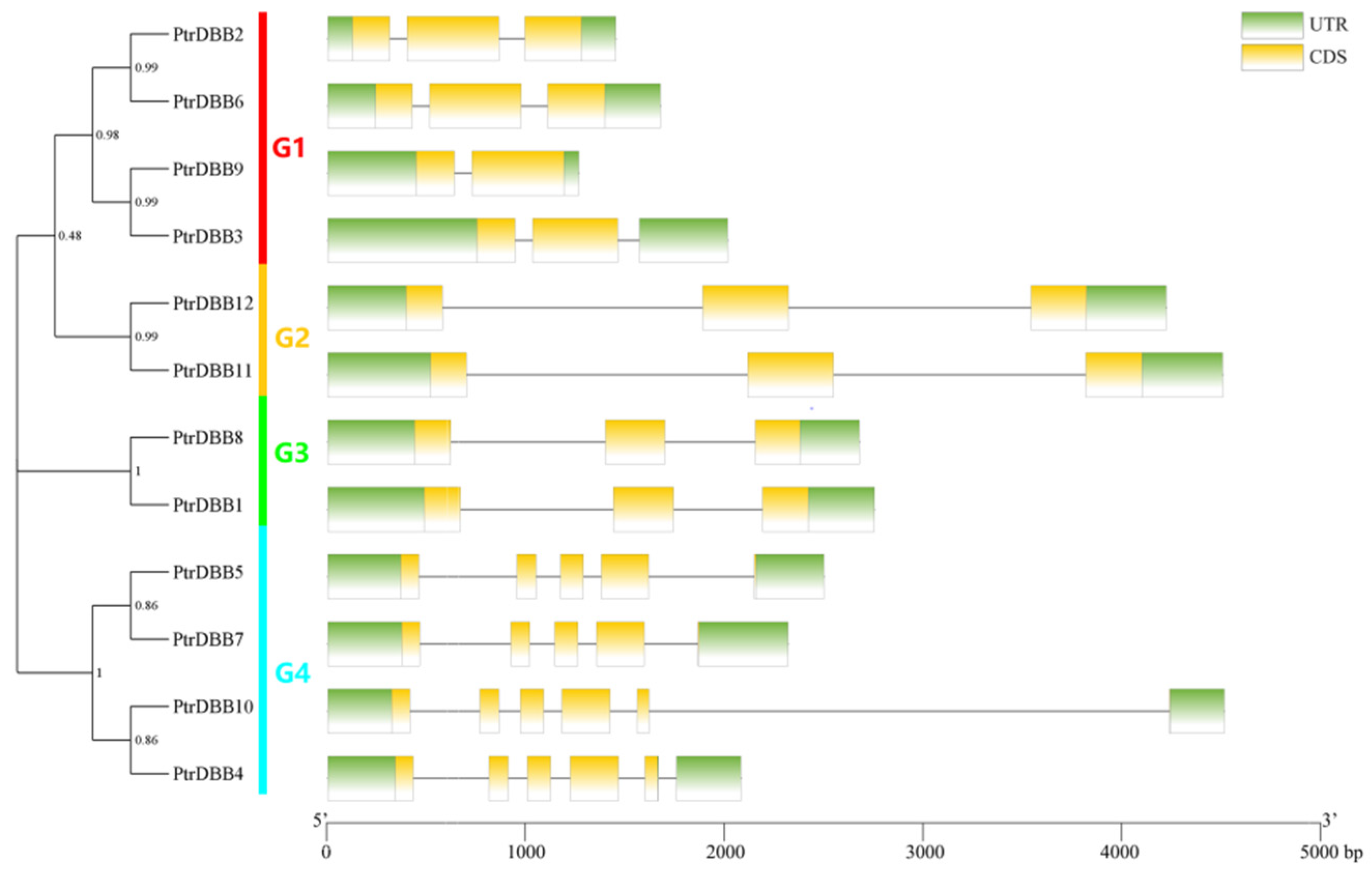
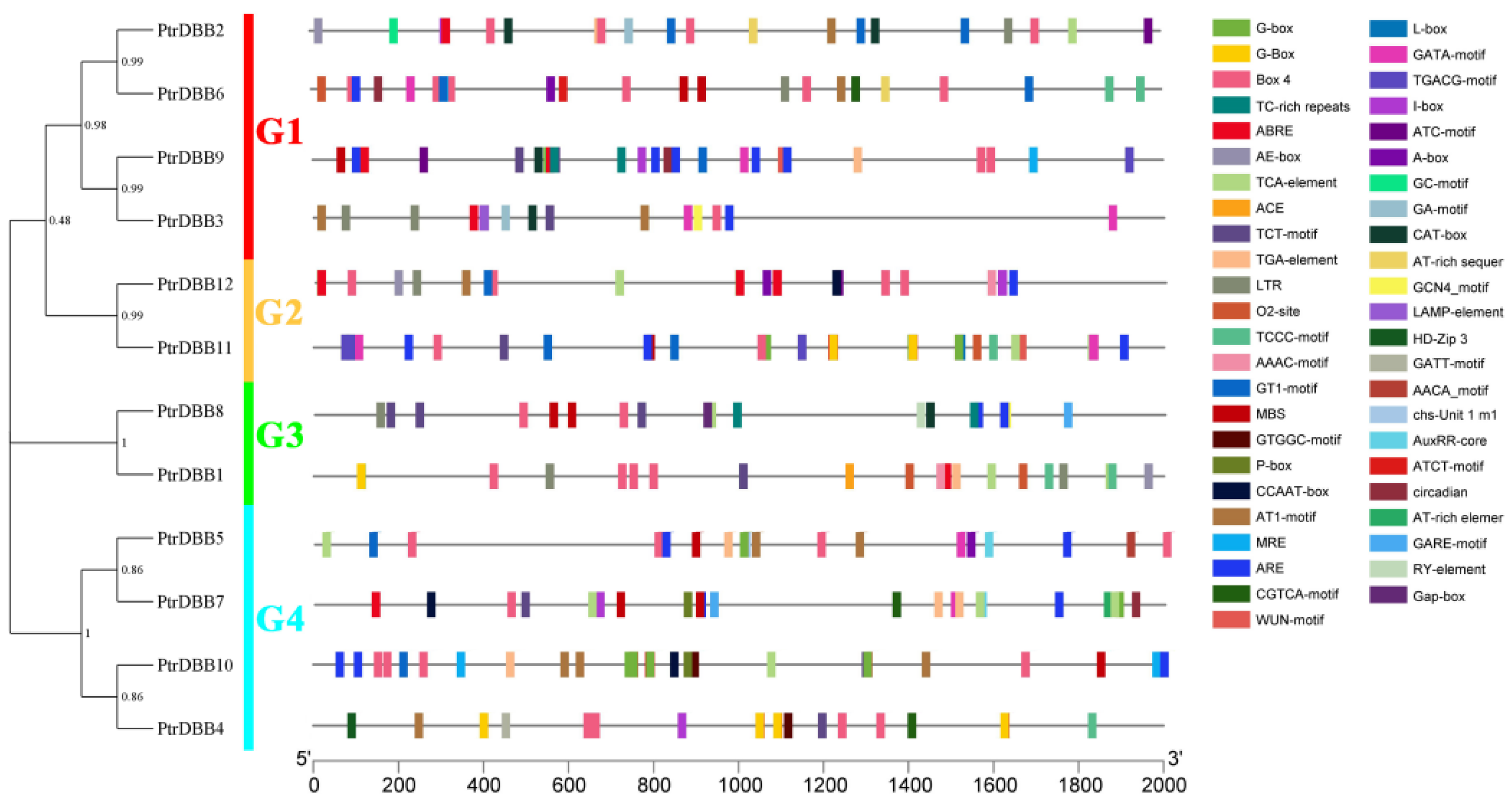
2.2. Analysis of Phylogenetic Relationship and Gene Structures of the DBB Genes
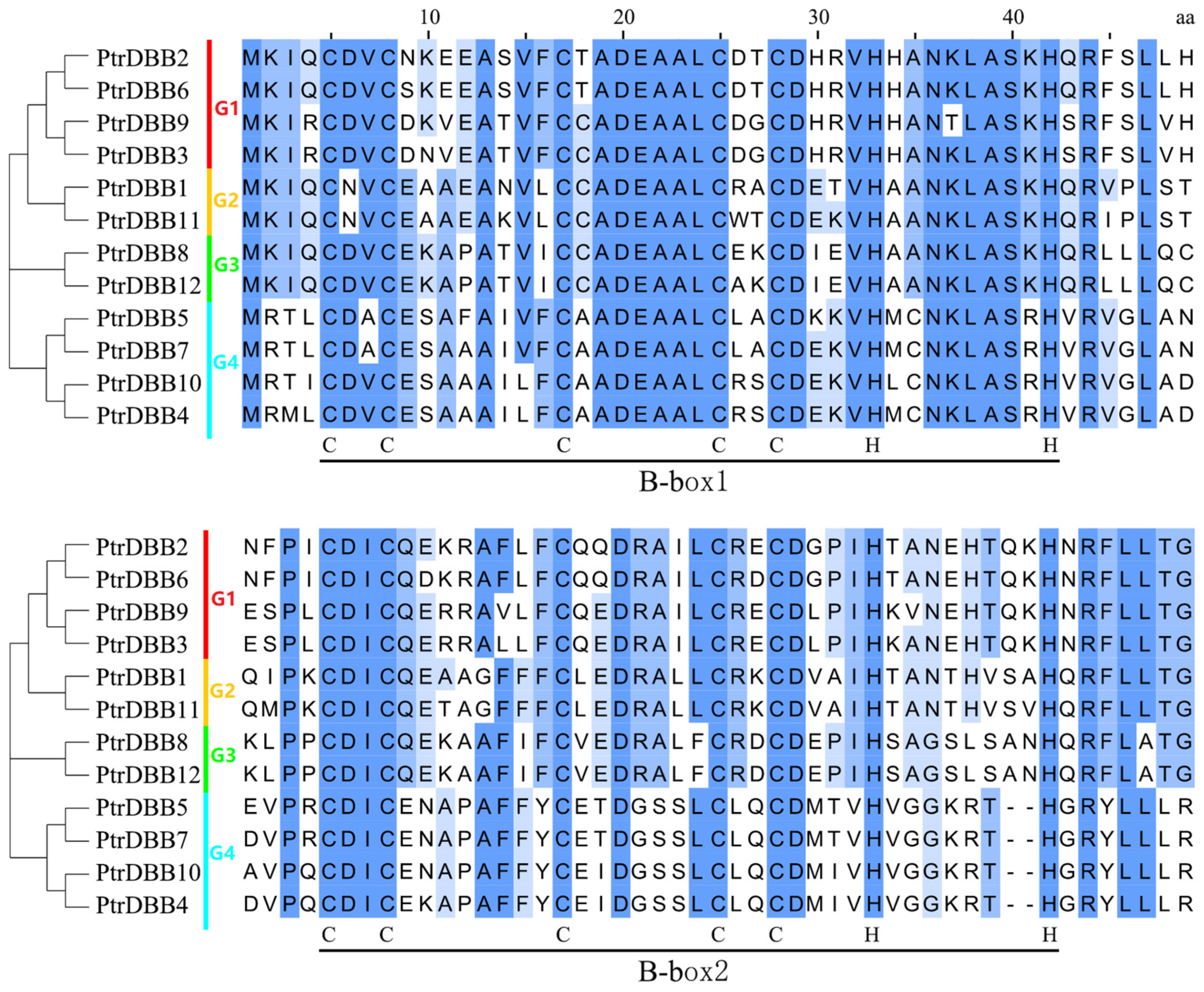
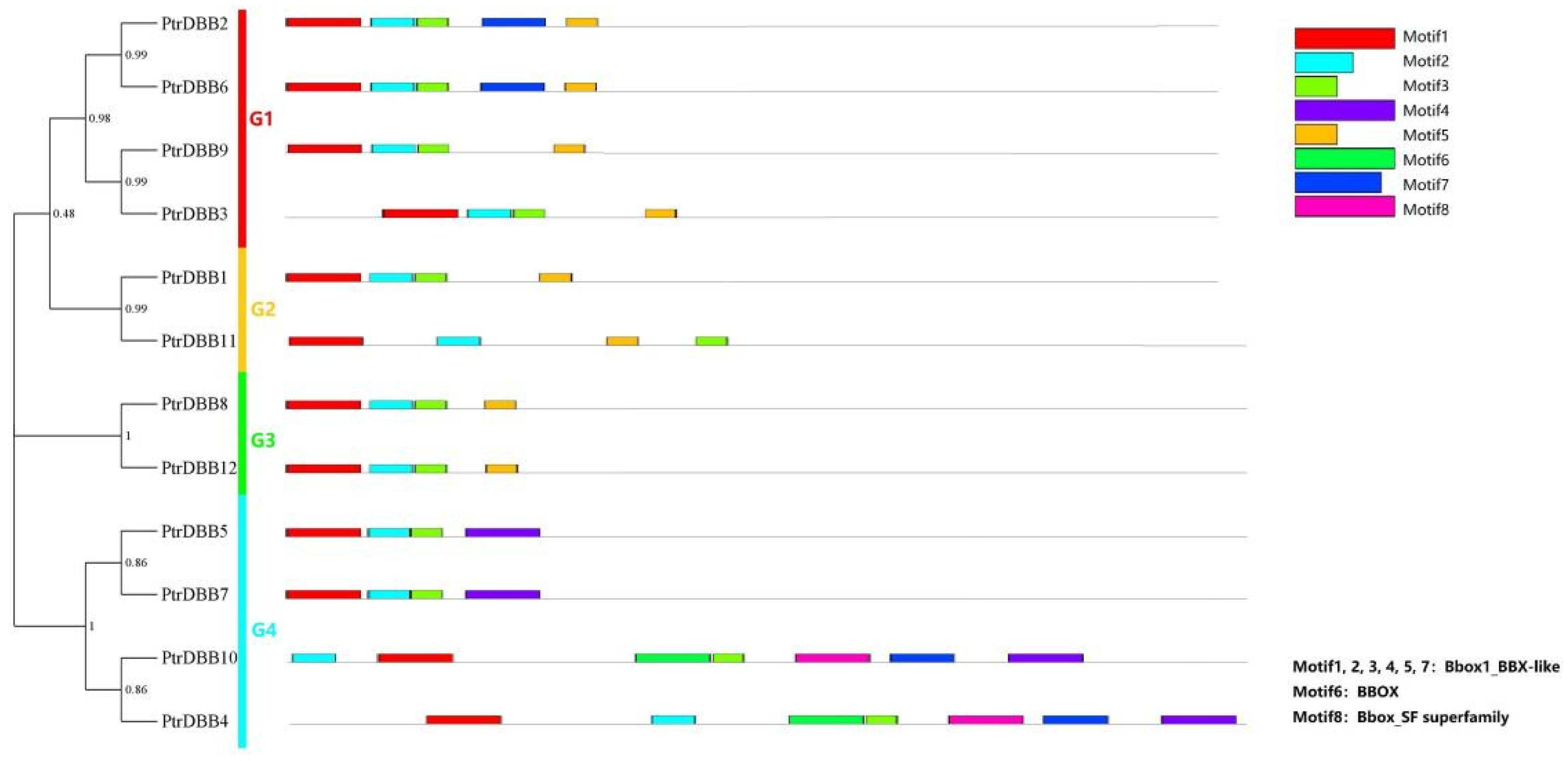
2.3. Analysis of Conservative Motif and Homology Modeling
2.4. Chromosomal Location and Gene Duplications of PtrDBBs
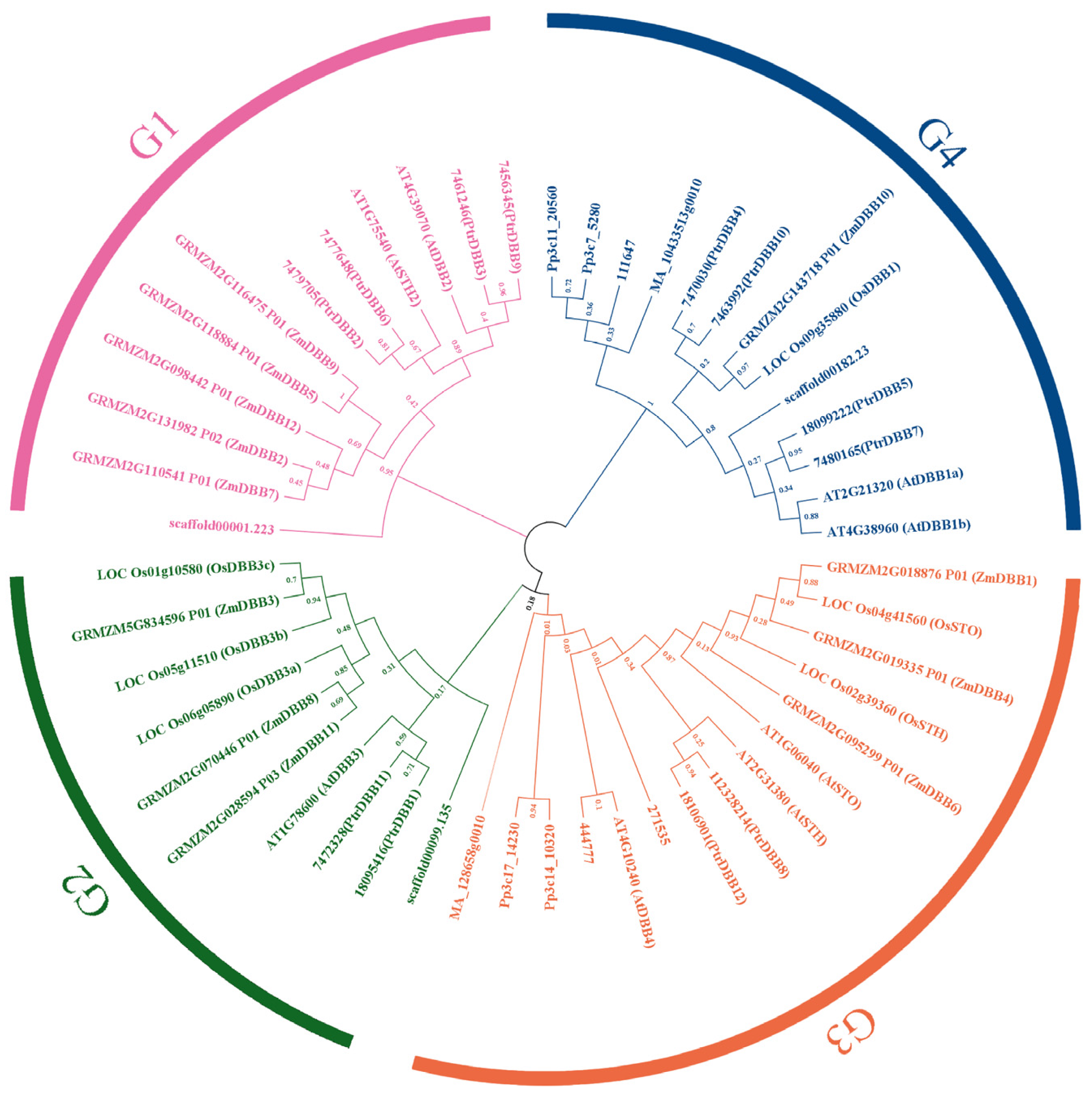
2.5. Evolution Profiles and Divergence of DBB Genes
2.6. Expression Patterns of DBB Genes in Populus trichocarpa
2.7. Cis-Regulatory Elements Analysis of PtrDBBs
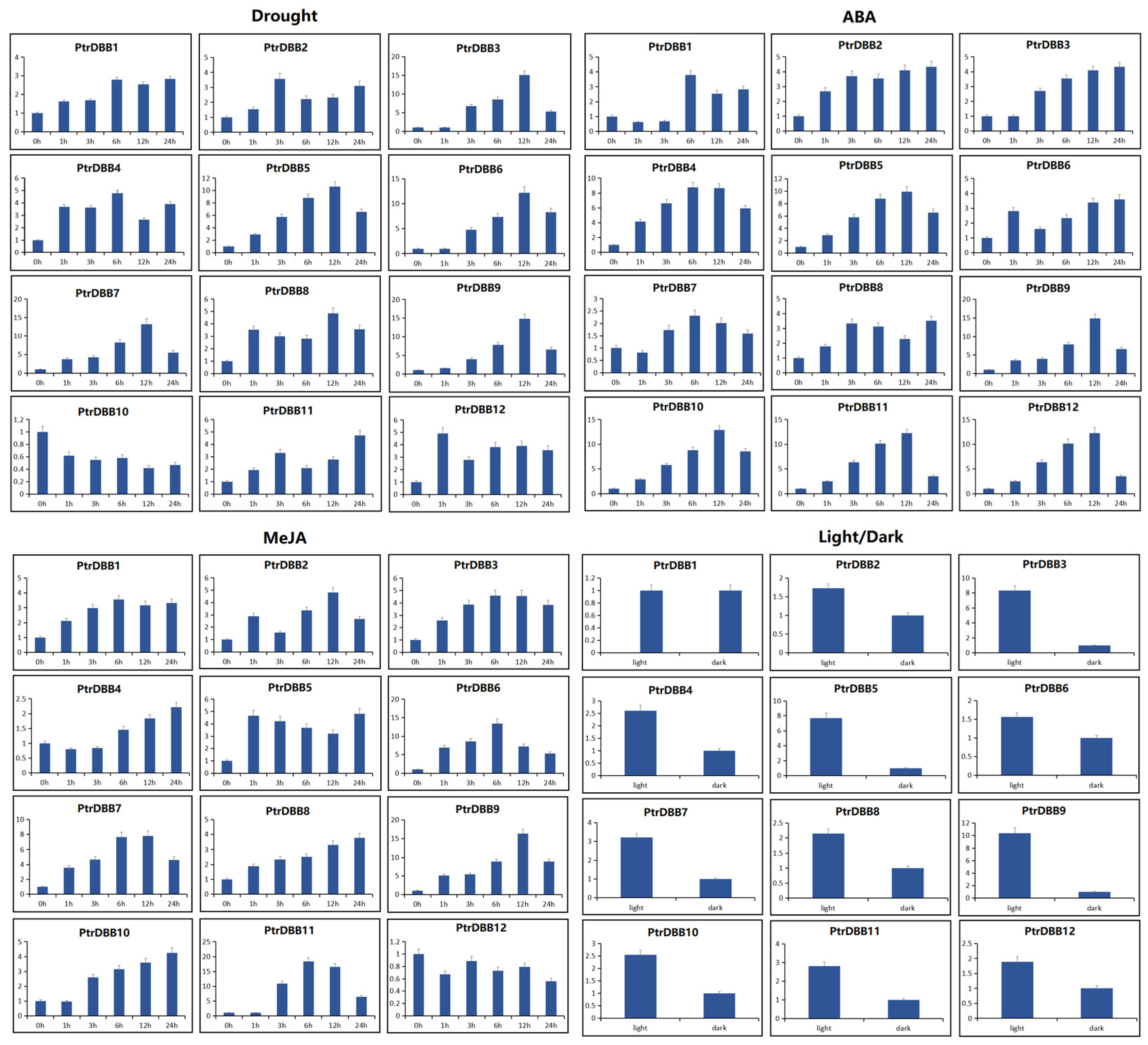
2.8. Expression Patterns of PtrDBB Genes under Abiotic Stress and Phytohormone Treatments
3. Discussion
3.1. PtrDBBs in P. trichocarpa
3.2. Evolutionary Patterns of DBBs in P. trichocarpa
3.3. Potential Roles of PtrDBBs in P. trichocarpa
3.4. Stress and Phytohormones Induced Expression of PtrDBBs in P. trichocarpa
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Identification of PtrDBB Genes in Populus and Phylogenetic Analysis
4.3. Motif Structure, Gene Structure, Protein Structure, and PtrDBB Gene Promoter Analysis
4.4. Chromosomal Location, Synteny Analysis and Duplication Events
4.5. PtrDBB Expression Profiles and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| BBX | B-box protein |
| CDD | conserved domain database |
| DBB | double B-box |
| DRE | osmotic-responsive element |
| LTRE | cold-responsive element |
| MeJA | methyl jasmonate |
| MW | molecular weight |
| MYA | million years ago |
| PEG | polyethylene glycol 6000 |
| pI | isoelectric point |
| Ptr | Populus trichocarpa |
| SA | silica acid |
| FM | female catkins, prior to seed release |
| F | female catkins, post-fertilization |
| M | male catkins |
| ML | mature leaf |
| REF | washed fibrous roots |
| RTC | roots from plants in tissue culture |
| AxB | axillary bud |
| YFB | newly initiated female floral buds |
| YMB | newly initiated male floral buds |
| Xylem1 | developing phloem |
| PC | phloem, cortex, epidermis |
| Phloem3 | developing phloem/cambium |
References
- Gong, W.; Shen, Y.P.; Ma, L.G.; Pan, Y.; Du, Y.L.; Wang, D.H.; Yang, J.Y.; Hu, L.D.; Liu, X.F.; Dong, C.X. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004, 135, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Wu, T.; Zhang, J.; Wang, Z.; Pei, T.; Yang, H.; Li, J.; Xu, X. Genome-wide analyses of the genetic screening of C2H2-type zinc finger transcription factors and abiotic and biotic stress responses in tomato (Solanum iycopersicum) based on RNA-Seq data. Front. Genet. 2020, 11, 540. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Holm, M.; Deng, X.W. The B-box domain protein BBX21 promotes Photomorphogenesis. Plant Physiol. 2018, 176, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Massiah, M.A.; Matts, J.A.; Short, K.M.; Simmons, B.N.; Singireddy, S.; Yi, Z.; Cox, T.C. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: Insights into an evolutionary conserved RING fold. J. Mol. Biol. 2007, 369, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, J.; Deng, K.; Tu, X.; Zhao, X.; Tang, D.; Liu, X. DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue lightmediated hypocotyl elongation in Arabidopsis. Planta 2011, 233, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Fan, S.; Jia, P.; Li, G.; Muhammad, I.; Li, Y.; Sharif, R.; Dong, F.; Zuo, X.; Li, K. Genome identification of B-BOX gene family members in seven rosaceae species and their expression analysis in response to flower induction in malus domestica. Molecules 2018, 23, 1763. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, T.; Ito, S.; Norihito, N.N.; Niwa, Y.; Murakami, M.; Yamashino, T.; Mizuno, T. The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian associated events in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2008, 72, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.S.; Li, Y.H.; Chen, L.T.; Chen, W.C.; Hsieh, W.P. LZF1, a HY5 regulated transcriptional factor, functions in Arabidopsis deetiolation. Plant J. 2008, 54, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Heng, Y.; Lin, F.; Jiang, Y.; Ding, M.; Yan, T.; Lan, H. B-box containing proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate Photo-morphogenesis in Arabidopsis. Plant Physiol. 2019, 180, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Heng, Y.; Wang, X.; Deng, X.W.; Xu, D. A positive feedback loop of BBX11-BBX21-HY5 promotes Photomorphogenic development in Arabidopsis. Plant Commun. 2020, 1, 100045. [Google Scholar]
- Jia, F.; Wu, B.; Li, H.; Huang, J.; Zheng, C. Genome-wide identification and characterization of F-box family in maize. Mol. Genet. Genom. 2013, 288, 559–577. [Google Scholar] [CrossRef]
- Li, W.L.; Jing, C.; Qi, S. Expression analysis of genes encoding double B-box zinc finger proteins in maize. Funct. Integr. Genomic 2017, 17, 653–666. [Google Scholar] [CrossRef]
- Yan, H.; Marquardt, K.; Indorf, M.; Jutt, D.; Kircher, S.; Neuhaus, G. Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 2011, 156, 1772–1782. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Liao, J.; Chen, Q.; Jin, W.; Li, S.; Zhu, T.; Li, S. Identification and characterization of the BBX gene family in Bambusa pervariabilis×Dendrocalamopsis grandis and their potential role under adverse environmental stresses. Int. J. Mol. Sci. 2023, 24, 13465. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Fu, D.; Gao, X.; Zeng, Q.; Chen, X.; Wu, J.; Zhang, N. Characterization and expression profiles of the B-box gene family during plant growth and under low-nitrogen stress in Saccharum. BMC Genom. 2023, 24, 79. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZBBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2020, 229, 2707–2729. [Google Scholar] [CrossRef] [PubMed]
- Soding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Dai, J.; Liu, X.; Lin, D. Genome-wide and expression analysis of B-box gene family in pepper. BMC Genom. 2021, 22, 883. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Yu, R.; Li, G. Identifcation and expression analysis of the B-box transcription factor family in pepper. Acta Hortic. Sinica 2021, 48, 1–15. [Google Scholar]
- Feng, Z.; Li, M.Y.; Li, Y. Comprehensive identification and expression analysis of B-Box genes in cotton. BMC Genom. 2021, 22, 439. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.N.; Wang, X.; Li, Y. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in Tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef]
- Wei, H.; Wang, P.; Chen, J.; Li, C.; Wang, Y.; Yuan, Y.; Fang, J.; Leng, X. Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol. 2020, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Wang, R.; Wang, R.; Yang, S.; Yang, Y. Genome-wide identiffcation, phylogenetic analysis, and expression proffling of the BBX family genes in pear. J. Hortic. Sci. Biotechnol. 2017, 93, 37–50. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 202, 8–37. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Johansson, H.; Hettiarachchi, C.; Irigoyen, M.L.; Desai, M. LZF1/SALTTOLERANCEHOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 2008, 20, 2324–2338. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.P.; Fan, G.Q.; Zhao, Z.L.; Deng, M.J. Compatible solute, transporter protein, transcription factor, and hormone-related gene expression provides an indicator of drought stress in Paulownia fortunei. Funct. Integr. Genom. 2014, 14, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef] [PubMed]
- Reiser, L.; Subramaniam, S.; Li, D.; Huala, E. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr. Protoc. Bioinform. 2017, 60, 1–11. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 427, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 3784, 3788. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van, D. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 325, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. Tbtools an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 1194, 1202. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 77, 80–88. [Google Scholar] [CrossRef]
- Rodgers-Melnick, M.; Mane, S.P.; Dharmawardhana, P.; Slavov, G.T.; Crasta, O.R.; Strauss, S.H.; Brunner, A.M.; DiFazio, S.P. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 2012, 22, 95–105. [Google Scholar] [CrossRef] [PubMed]
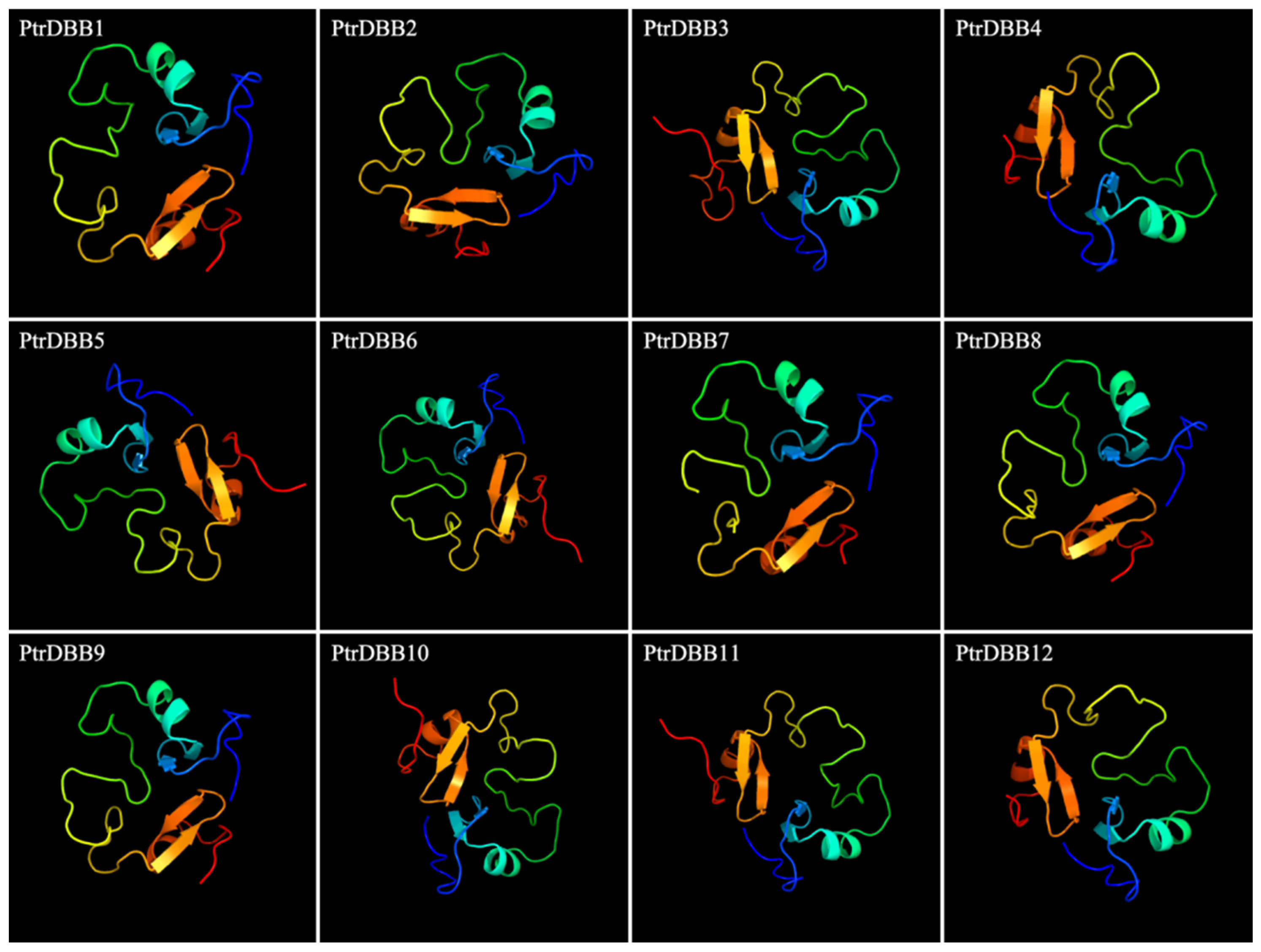
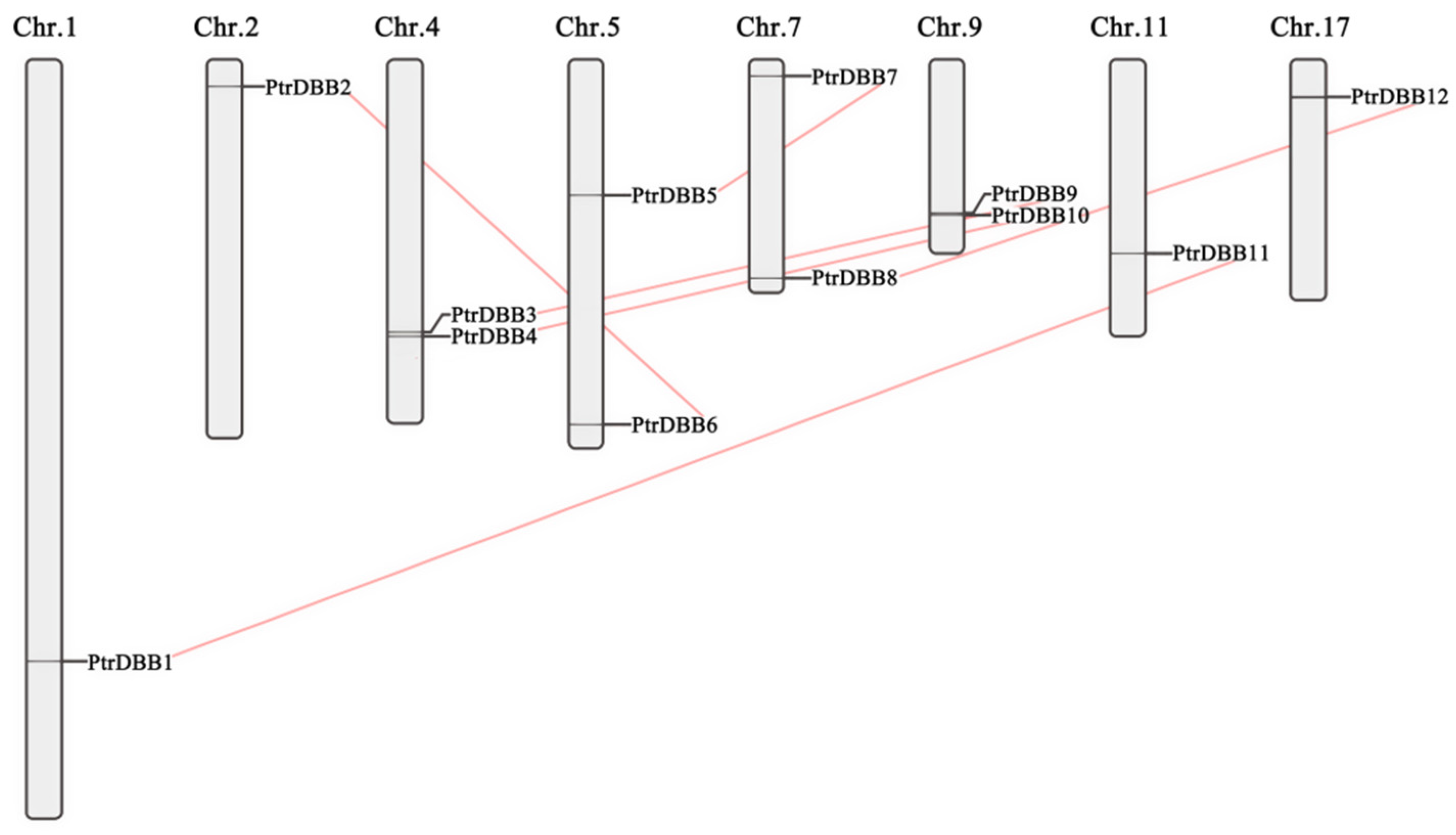
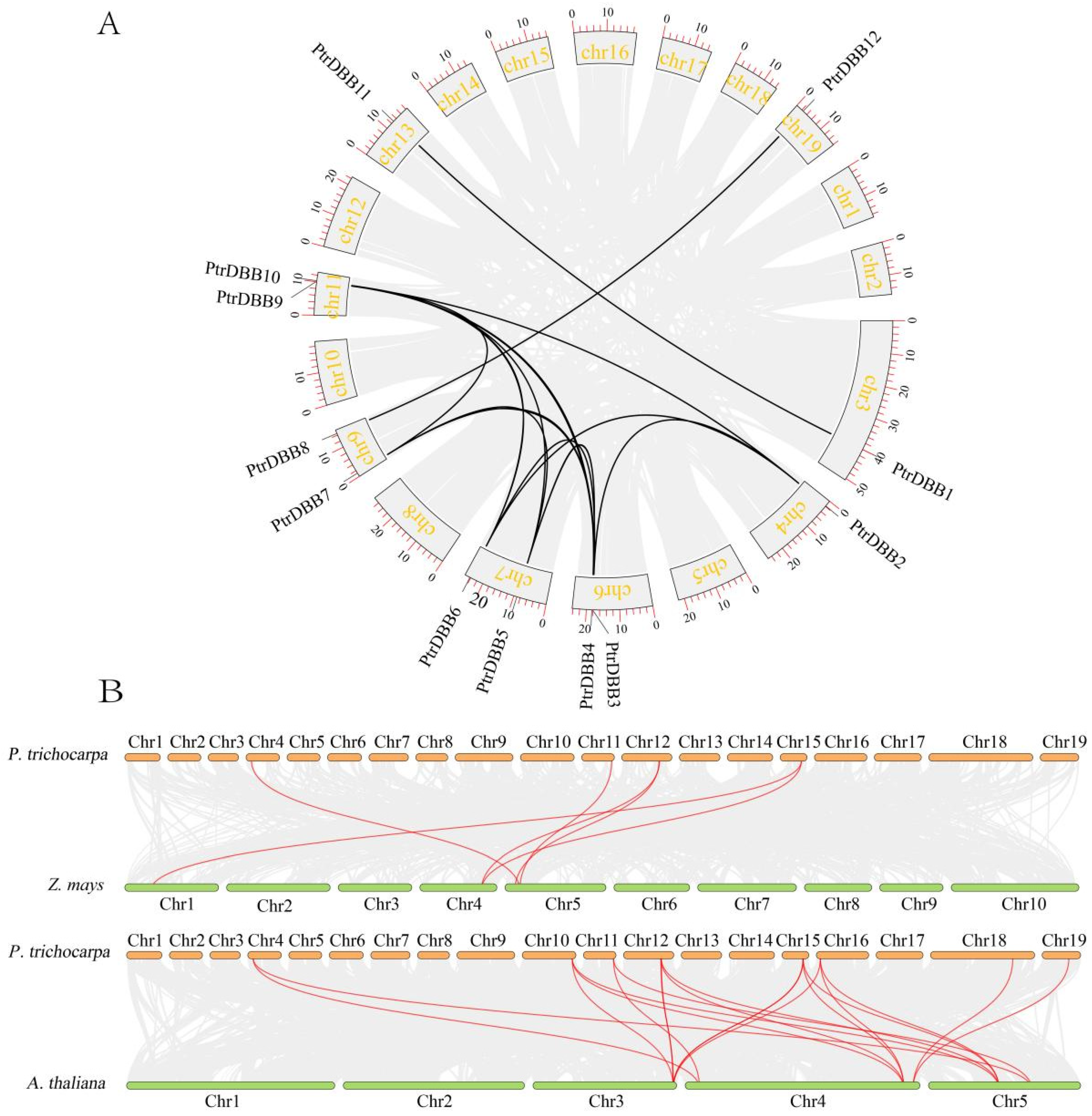

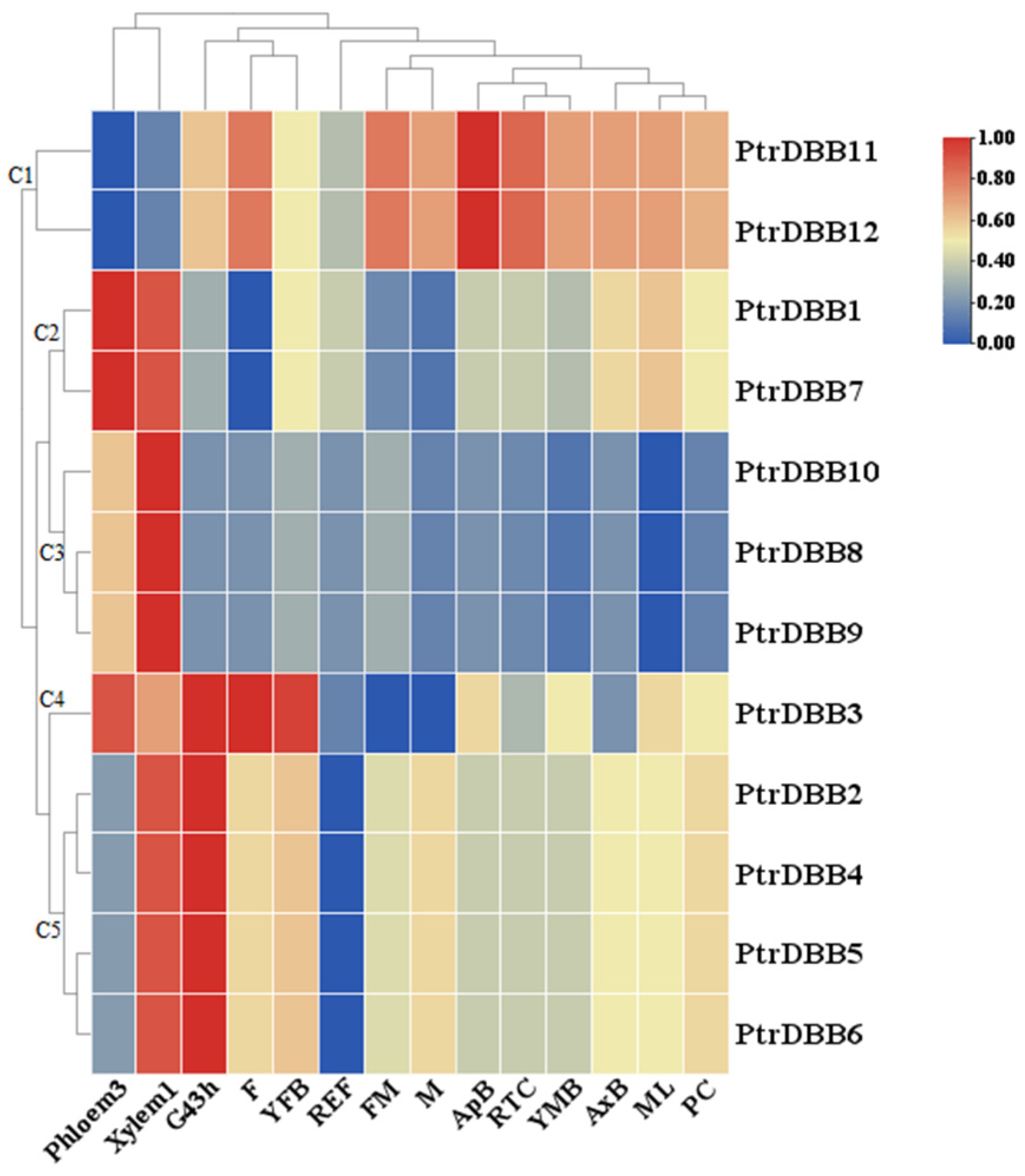
| Gene Name | Gene ID | Location | CDS Length (bp) | Size (aa) | Protein Mw (Da) | pI | Exons |
|---|---|---|---|---|---|---|---|
| PtrDBB1 | 18095416 | 1:39,971,068–39,975,286(+) | 894 | 298 | 31,852.75 | 5.75 | 3 |
| PtrDBB2 | 7479705 | 2:1,813,398–1,814,847(−) | 933 | 311 | 34,336.37 | 5.79 | 3 |
| PtrDBB3 | 7461246 | 4:18,137,086–18,139,098(+) | 621 | 207 | 22,859.31 | 5.72 | 2 |
| PtrDBB4 | 7470030 | 4:18,368,982–18,371,060(−) | 612 | 204 | 22,668.51 | 5.81 | 5 |
| PtrDBB5 | 18099222 | 5:9,032,208–9,034,704(+) | 555 | 185 | 20,506.21 | 7.05 | 5 |
| PtrDBB6 | 7477648 | 5:24,240,055–24,241,728(+) | 936 | 312 | 34,288.43 | 6.20 | 3 |
| PtrDBB7 | 7480165 | 7:1,136,453–1,138,769(+) | 558 | 186 | 20,515.17 | 6.23 | 5 |
| PtrDBB8 | 112328214 | 7:14,516,505–14,519,179(−) | 708 | 236 | 26,022.39 | 4.80 | 3 |
| PtrDBB9 | 7456345 | 9:10,201,380–10,202,642(+) | 654 | 218 | 24,021.86 | 6.39 | 2 |
| PtrDBB10 | 7463992 | 9:10,345,821–10,350,331(−) | 612 | 204 | 22,650.42 | 6.24 | 6 |
| PtrDBB11 | 7472328 | 11:12,884,783–12,889,285(+) | 897 | 299 | 32,284.26 | 6.00 | 3 |
| PtrDBB12 | 18106901 | 17:2,528,459–2,531,209(+) | 717 | 239 | 25,989.39 | 4.77 | 3 |
| Duplicate Gene Pair | Ka | Ks | Ka/Ks | Purify Selection | Duplication Type | Time (MYA a) |
|---|---|---|---|---|---|---|
| PtrDBB1/PtrDBB9 | 0.540005 | 2.643361 | 0.204287 | YES | Segmental | 203.3354662 |
| PtrDBB1/PtrDBB10 | 0.800996 | 2.706739 | 0.295927 | YES | Segmental | 208.2107045 |
| PtrDBB1/PtrDBB11 | 0.069298 | 0.231111 | 0.299846 | YES | Segmental | 17.77776362 |
| PtrDBB1/PtrDBB3 | 0.700382 | 2.136888 | 0.327758 | YES | Segmental | 164.3760115 |
| PtrDBB1/PtrDBB6 | 0.657927 | 1.818373 | 0.361822 | YES | Segmental | 139.874857 |
| PtrDBB2/PtrDBB9 | 0.280933 | 2.721383 | 0.103232 | YES | Segmental | 209.3371921 |
| PtrDBB2/PtrDBB6 | 0.052123 | 0.325995 | 0.159889 | YES | Segmental | 25.07652646 |
| PtrDBB2/PtrDBB12 | 0.569719 | 2.558617 | 0.222667 | YES | Segmental | 196.816677 |
| PtrDBB2/PtrDBB11 | 0.669162 | 2.946344 | 0.227116 | YES | Segmental | 226.6418438 |
| PtrDBB2/PtrDBB8 | 0.505474 | 1.951101 | 0.259071 | YES | Segmental | 150.0846888 |
| PtrDBB4/PtrDBB7 | 0.192639 | 1.176669 | 0.163715 | YES | Segmental | 90.51298954 |
| PtrDBB5/PtrDBB10 | 0.189565 | 1.388261 | 0.136549 | YES | Segmental | 106.7893365 |
| PtrDBB5/PtrDBB4 | 0.206347 | 1.115763 | 0.184938 | YES | Segmental | 85.82789923 |
| PtrDBB5/PtrDBB7 | 0.074573 | 0.267214 | 0.279075 | YES | Segmental | 20.55494785 |
| PtrDBB6/PtrDBB9 | 0.319537 | 2.137573 | 0.149486 | YES | Segmental | 164.4287226 |
| PtrDBB6/PtrDBB12 | 0.609082 | 2.298195 | 0.265026 | YES | Segmental | 176.7842262 |
| PtrDBB6/PtrDBB11 | 0.714865 | 2.282156 | 0.313241 | YES | Segmental | 175.5504411 |
| PtrDBB7/PtrDBB10 | 0.189482 | 1.393803 | 0.135946 | YES | Segmental | 107.2155891 |
| PtrDBB8/PtrDBB12 | 0.058761 | 0.256519 | 0.22907 | YES | Segmental | 19.73223654 |
| PtrDBB8/PtrDBB6 | 0.514803 | 1.787578 | 0.287989 | YES | Segmental | 137.5059917 |
| PtrDBB9/PtrDBB11 | 0.48228 | 2.291287 | 0.210484 | YES | Segmental | 176.2528183 |
| PtrDBB9/PtrDBB12 | 0.649449 | 2.427373 | 0.267552 | YES | Segmental | 186.7210054 |
| PtrDBB10/PtrDBB1 | 0.819611 | 3.002887 | 0.272941 | YES | Segmental | 230.9912805 |
| PtrDBB11/PtrDBB12 | 0.544453 | 4.85965 | 0.112035 | YES | Segmental | 373.8192222 |
| PtrDBB11/PtrDBB2 | 0.638452 | 3.029696 | 0.210731 | YES | Segmental | 233.0535028 |
| PtrDBB11/PtrDBB7 | 0.671339 | 1.771357 | 0.378997 | YES | Segmental | 136.2582048 |
| PtrDBB12/PtrDBB7 | 0.694002 | 3.017405 | 0.23 | YES | Segmental | 232.1080693 |
| Duplicate Gene Pair | Ka | Ks | Ka/Ks | Purify Selection | Duplication Type | Time (MYA a) |
|---|---|---|---|---|---|---|
| PtrDBB1/AT1G78600 | 0.273673 | 1.70025 | 0.160960704 | YES | Segmental | 130.7884638 |
| PtrDBB1/AT1G78600 | 0.273673 | 1.750689 | 0.15632326 | YES | Segmental | 134.6684 |
| PtrDBB2/AT4G38960 | 0.673689 | 1.795728 | 0.375162313 | YES | Segmental | 138.1329048 |
| PtrDBB2/AT2G31380 | 0.568092 | 2.702995 | 0.210171206 | YES | Segmental | 207.9227005 |
| PtrDBB4/AT4G38960 | 0.224073 | 2.133469 | 0.105027385 | YES | Segmental | 164.1130232 |
| PtrDBB4/AT2G21320 | 0.231998 | 2.386342 | 0.097219099 | YES | Segmental | 183.5647708 |
| PtrDBB4/AT1G78600 | 0.774086 | 3.386257 | 0.228596496 | YES | Segmental | 260.4812822 |
| PtrDBB5/AT1G78600 | 0.694743 | 4.550277 | 0.152681432 | YES | Segmental | 350.0212923 |
| PtrDBB6/AT4G38960 | 0.729036 | 2.007423 | 0.363170212 | YES | Segmental | 154.4171159 |
| PtrDBB6/AT2G31380 | 0.599408 | 3.301109 | 0.181577634 | YES | Segmental | 253.9314842 |
| PtrDBB7/AT1G78600 | 0.639815 | 1.956903 | 0.326952767 | YES | Segmental | 150.5310212 |
| PtrDBB9/AT4G39070 | 0.350527 | 1.938564 | 0.180817944 | YES | Segmental | 149.1203068 |
| PtrDBB10/AT4G38960 | 0.174062 | 2.238844 | 0.077746495 | YES | Segmental | 172.2187941 |
| PtrDBB11/AT1G78600 | 0.279831 | 1.596113 | 0.175320488 | YES | Segmental | 122.7779101 |
| PtrDBB11/AT1G78600 | 0.300215 | 1.791754 | 0.167553531 | YES | Segmental | 137.8272228 |
| PtrDBB11/AT4G39070 | 0.538742 | 2.51422 | 0.214277967 | YES | Segmental | 193.4015524 |
| PtrDBB11/AT2G24790 | 0.914811 | 3.624656 | 0.252385571 | YES | Segmental | 278.819697 |
| PtrDBB12/AT4G38960 | 0.728341 | 1.671692 | 0.435690773 | YES | Segmental | 128.5917088 |
| PtrDBB12/AT2G31380 | 0.242356 | 3.907907 | 0.062016731 | YES | Segmental | 300.6082636 |
| PtrDBB1/GRMZM2G422644 | 0.724478 | 3.282537 | 0.220707 | YES | Segmental | 252.5028 |
| PtrDBB1/GRMZM2G143718 | 0.687263 | 3.173714 | 0.216549 | YES | Segmental | 244.1319 |
| PtrDBB1/GRMZM2G028594 | 0.398684 | 3.647221 | 0.109312 | YES | Segmental | 280.5554 |
| PtrDBB2/GRMZM2G075562 | 1.504925 | 2.630359 | 0.572137 | YES | Segmental | 202.3353 |
| PtrDBB2/GRMZM2G028594 | 1.001651 | 2.3692 | 0.422781 | YES | Segmental | 182.2461 |
| PtrDBB2/GRMZM2G422644 | 0.716321 | 2.35668 | 0.303953 | YES | Segmental | 181.2831 |
| PtrDBB2/GRMZM2G095299 | 0.562875 | 2.550898 | 0.220658 | YES | Segmental | 196.2229 |
| PtrDBB4/GRMZM2G098442 | 0.624664 | 3.24372 | 0.192576 | YES | Segmental | 249.5169 |
| PtrDBB4/GRMZM2G143718 | 0.2927 | 1.754125 | 0.166864 | YES | Segmental | 134.9327 |
| PtrDBB4/GRMZM2G422644 | 0.297375 | 2.10252 | 0.141437 | YES | Segmental | 161.7323 |
| PtrDBB6/GRMZM2G075562 | 1.09016 | 2.779934 | 0.392153 | YES | Segmental | 213.8411 |
| PtrDBB6/GRMZM2G095299 | 0.675375 | 2.269925 | 0.297532 | YES | Segmental | 174.6096 |
| PtrDBB7/GRMZM2G028594 | 1.055774 | 2.892911 | 0.364952 | YES | Segmental | 222.5316 |
| PtrDBB8/GRMZM2G028594 | 1.014354 | 1.755839 | 0.577703 | YES | Segmental | 135.0645 |
| PtrDBB8/GRMZM2G075562 | 1.107168 | 2.349007 | 0.471335 | YES | Segmental | 180.6929 |
| PtrDBB10/GRMZM2G098442 | 0.6718 | 2.342246 | 0.286819 | YES | Segmental | 180.1727 |
| PtrDBB10/GRMZM2G422644 | 0.279454 | 1.801646 | 0.15511 | YES | Segmental | 138.5881 |
| PtrDBB10/GRMZM2G143718 | 0.255699 | 1.770641 | 0.144411 | YES | Segmental | 136.2031 |
| PtrDBB11/GRMZM2G143718 | 0.663161 | 2.435664 | 0.272271 | YES | Segmental | 187.3587 |
| PtrDBB11/GRMZM2G028594 | 0.462405 | 3.441831 | 0.134348 | YES | Segmental | 264.7562 |
| PtrDBB12/GRMZM2G028594 | 0.840264 | 2.622381 | 0.32042 | YES | Segmental | 201.7216 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, R.; Li, Y.; Wang, L.; Li, Z.; Wu, R.; Xu, K.; Liu, Y. The DBB Family in Populus trichocarpa: Identification, Characterization, Evolution and Expression Profiles. Molecules 2024, 29, 1823. https://doi.org/10.3390/molecules29081823
Wu R, Li Y, Wang L, Li Z, Wu R, Xu K, Liu Y. The DBB Family in Populus trichocarpa: Identification, Characterization, Evolution and Expression Profiles. Molecules. 2024; 29(8):1823. https://doi.org/10.3390/molecules29081823
Chicago/Turabian StyleWu, Ruihua, Yuxin Li, Lin Wang, Zitian Li, Runbin Wu, Kehang Xu, and Yixin Liu. 2024. "The DBB Family in Populus trichocarpa: Identification, Characterization, Evolution and Expression Profiles" Molecules 29, no. 8: 1823. https://doi.org/10.3390/molecules29081823
APA StyleWu, R., Li, Y., Wang, L., Li, Z., Wu, R., Xu, K., & Liu, Y. (2024). The DBB Family in Populus trichocarpa: Identification, Characterization, Evolution and Expression Profiles. Molecules, 29(8), 1823. https://doi.org/10.3390/molecules29081823





