Effects of Red Sorghum-Derived Deoxyanthocyanidins and Their O-β-D-Glucosides on E-Cadherin Promoter Activity in PC-3 Prostate Cancer Cells
Abstract
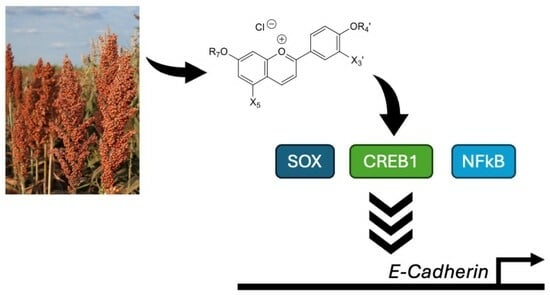
1. Introduction
2. Results
2.1. Effects of Synthesized Pigments on Cell Viability
2.2. Influences of Synthesized Pigments on the Expression of E-Cadherin-Related Genes
2.3. Physicochemical Properties and Drug-likeness Prediction
3. Discussion
4. Materials and Methods
4.1. Synthesis of Compounds
4.2. Reagents
4.3. Cell Culture
4.4. Cell Viability Assay
4.5. Plasmids and Transfection
4.6. In Silico Physicochemical Properties and Drug-likeness Evaluation
4.7. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yoshida, K.; Mori, M.; Kondo, T. Blue Flower Color Development by Anthocyanins: From Chemical Structure to Cell Physiology. Nat. Prod. Rep. 2009, 26, 884–915. [Google Scholar] [CrossRef] [PubMed]
- Galland, S.; Mora, N.; Abert-Vian, M.; Rakotomanomana, N.; Dangles, O. Chemical Synthesis of Hydroxycinnamic Acid Glucosides and Evaluation of Their Ability to Stabilize Natural Colors via Anthocyanin Copigmentation. J. Agric. Food Chem. 2007, 55, 7573–7579. [Google Scholar] [CrossRef] [PubMed]
- Willstätter, R.; Everest, A.E. Untersuchungen Über Die Anthocyane. I. Über Den Farbstoff Der Kornblume. Justus Liebigs Ann. Chem. 1913, 401, 189–232. [Google Scholar] [CrossRef]
- Asen, S.; Stewart, R.N.; Norris, K.H. Anthocyanin, Flavonol Copigments, and pH Responsible for Larkspur Flower Color. Phytochemistry 1975, 14, 2677–2682. [Google Scholar] [CrossRef]
- Tsuda, T. Dietary Anthocyanin-Rich Plants: Biochemical Basis and Recent Progress in Health Benefits Studies. Mol. Nutr. Food Res. 2012, 56, 159–170. [Google Scholar] [CrossRef]
- Awika, J.M.; Rooney, L.W.; Waniska, R.D. Anthocyanins from Black Sorghum and Their Antioxidant Properties. Food Chem. 2005, 90, 293–301. [Google Scholar] [CrossRef]
- Mazza, G.; Brouillard, R. Color Stability and Structural Transformations of Cyanidin 3,5-Diglucoside and Four 3-Deoxyanthocyanins in Aqueous Solutions. J. Agric. Food Chem. 1987, 35, 422–426. [Google Scholar] [CrossRef]
- Pina, F.; Melo, M.J.; Laia, C.A.; Parola, A.J.; Lima, J.C. Chemistry and Applications of Flavylium Compounds: A Handful of Colours. Chem. Soc. Rev. 2012, 41, 869–908. [Google Scholar] [CrossRef]
- Yang, L.; Dykes, L.; Awika, J.M. Thermal Stability of 3-Deoxyanthocyanidin Pigments. Food Chem. 2014, 160, 246–254. [Google Scholar] [CrossRef]
- Awika, J.M.; Rooney, L.W. Sorghum Phytochemicals and Their Potential Impact on Human Health. Phytochemistry 2004, 65, 1199–1221. [Google Scholar] [CrossRef]
- Carbonneau, M.-A.; Cisse, M.; Mora-Soumille, N.; Dairi, S.; Rosa, M.; Michel, F.; Lauret, C.; Cristol, J.-P.; Dangles, O. Antioxidant Properties of 3-Deoxyanthocyanidins and Polyphenolic Extracts from Côte d’Ivoire’s Red and White Sorghums Assessed by ORAC and in Vitro LDL Oxidisability Tests. Food Chem. 2014, 145, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.R.; Belton, P.S.; Beta, T.; Duodu, K.G. Increasing the Utilisation of Sorghum, Millets and Pseudocereals: Developments in the Science of Their Phenolic Phytochemicals, Biofortification and Protein Functionality. J. Cereal Sci. 2014, 59, 257–275. [Google Scholar] [CrossRef]
- Al Bittar, S.; Mora, N.; Loonis, M.; Dangles, O. A Simple Synthesis of 3-Deoxyanthocyanidins and Their O-Glucosides. Tetrahedron 2016, 72, 4294–4302. [Google Scholar] [CrossRef]
- Mora-Soumille, N.; Al Bittar, S.; Rosa, M.; Dangles, O. Analogs of Anthocyanins with a 3′, 4′-Dihydroxy Substitution: Synthesis and Investigation of Their Acid–Base, Hydration, Metal Binding and Hydrogen-Donating Properties in Aqueous Solution. Dye. Pigment. 2013, 96, 7–15. [Google Scholar] [CrossRef]
- Bushweller, J.H. Targeting Transcription Factors in Cancer—From Undruggable to Reality. Nat. Rev. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a Selective Catalytic P300/CBP Inhibitor That Targets Lineage-Specific Tumours. Nature 2017, 550, 128–132. [Google Scholar] [CrossRef]
- Grimm, D.; Bauer, J.; Wise, P.; Krüger, M.; Simonsen, U.; Wehland, M.; Infanger, M.; Corydon, T.J. The Role of SOX Family Members in Solid Tumours and Metastasis. Semin. Cancer Biol. 2020, 67, 122–153. [Google Scholar] [CrossRef]
- Thomas-Jardin, S.E.; Dahl, H.; Nawas, A.F.; Bautista, M.; Delk, N.A. NF-κB Signaling Promotes Castration-Resistant Prostate Cancer Initiation and Progression. Pharmacol. Ther. 2020, 211, 107538. [Google Scholar] [CrossRef]
- Yagi, T.; Takeichi, M. Cadherin Superfamily Genes: Functions, Genomic Organization, and Neurologic Diversity. Genes. Dev. 2000, 14, 1169–1180. [Google Scholar] [CrossRef]
- Tran, N.L.; Nagle, R.B.; Cress, A.E.; Heimark, R.L. N-Cadherin Expression in Human Prostate Carcinoma Cell Lines. An Epithelial-Mesenchymal Transformation Mediating Adhesion withStromal Cells. Am. J. Pathol. 1999, 155, 787–798. [Google Scholar] [CrossRef]
- Birchmeier, W.; Behrens, J. Cadherin Expression in Carcinomas: Role in the Formation of Cell Junctions and the Prevention of Invasiveness. Biochim. Biophys. Acta 1994, 1198, 11–26. [Google Scholar] [CrossRef]
- Luo, J.; Lubaroff, D.M.; Hendrix, M.J. Suppression of Prostate Cancer Invasive Potential and Matrix Metalloproteinase Activity by E-Cadherin Transfection. Cancer Res. 1999, 59, 3552–3556. [Google Scholar] [PubMed]
- Pattabiraman, D.R.; Bierie, B.; Kober, K.I.; Thiru, P.; Krall, J.A.; Zill, C.; Reinhardt, F.; Tam, W.L.; Weinberg, R.A. Activation of PKA Leads to Mesenchymal-to-Epithelial Transition and Loss of Tumor-Initiating Ability. Science 2016, 351, aad3680. [Google Scholar] [CrossRef] [PubMed]
- Min, C.; Eddy, S.F.; Sherr, D.H.; Sonenshein, G.E. NF-kappaB and Epithelial to Mesenchymal Transition of Cancer. J. Cell Biochem. 2008, 104, 733–744. [Google Scholar] [CrossRef]
- Wegner, M. All Purpose Sox: The Many Roles of Sox Proteins in Gene Expression. Int. J. Biochem. Cell Biol. 2010, 42, 381–390. [Google Scholar] [CrossRef]
- Moreno, P.; Fexova, S.; George, N.; Manning, J.R.; Miao, Z.; Mohammed, S.; Muñoz-Pomer, A.; Fullgrabe, A.; Bi, Y.; Bush, N.; et al. Expression Atlas Update: Gene and Protein Expression in Multiple Species. Nucleic Acids Res. 2022, 50, D129–D140. [Google Scholar] [CrossRef] [PubMed]
- Chuang, K.-T.; Chiou, S.-S.; Hsu, S.-H. Recent Advances in Transcription Factors Biomarkers and Targeted Therapies Focusing on Epithelial-Mesenchymal Transition. Cancers 2023, 15, 3338. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Montanari, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Malzone, M.G.; Vanacore, D.; Di Franco, R.; La Mantia, E.; Iovane, G.; Piscitelli, R.; et al. Epithelial-Mesenchymal Transition in Prostate Cancer: An Overview. Oncotarget 2017, 8, 35376–35389. [Google Scholar] [CrossRef]
- Li, W.; Wu, D.; Niu, Z.; Jiang, D.; Ma, H.; He, H.; Zuo, X.; Xie, X.; He, Y. 5-Azacytidine Suppresses EC9706 Cell Proliferation and Metastasis by Upregulating the Expression of SOX17 and CDH1. Int. J. Mol. Med. 2016, 38, 1047–1054. [Google Scholar] [CrossRef]
- Du, Z.; Li, L.; Sun, W.; Zhu, P.; Cheng, S.; Yang, X.; Luo, C.; Yu, X.; Wu, X. Systematic Evaluation for the Influences of the SOX17/Notch Receptor Family Members on Reversing Enzalutamide Resistance in Castration-Resistant Prostate Cancer Cells. Front. Oncol. 2021, 11, 607291. [Google Scholar] [CrossRef]
- Staal, J.; Beyaert, R. Inflammation and NF-κB Signaling in Prostate Cancer: Mechanisms and Clinical Implications. Cells 2018, 7, 122. [Google Scholar] [CrossRef]
- Bishop, J.L.; Davies, A.; Ketola, K.; Zoubeidi, A. Regulation of Tumor Cell Plasticity by the Androgen Receptor in Prostate Cancer. Endocr. Relat. Cancer 2015, 22, R165–R182. [Google Scholar] [CrossRef]
- Shih, C.-H.; Siu, S.-O.; Ng, R.; Wong, E.; Chiu, L.C.M.; Chu, I.K.; Lo, C. Quantitative Analysis of Anticancer 3-Deoxyanthocyanidins in Infected Sorghum Seedlings. J. Agric. Food Chem. 2007, 55, 254–259. [Google Scholar] [CrossRef]
- Owumi, S.E.; Kazeem, A.I.; Wu, B.; Ishokare, L.O.; Arunsi, U.O.; Oyelere, A.K. Apigeninidin-Rich Sorghum Bicolor (L. Moench) Extracts Suppress A549 Cells Proliferation and Ameliorate Toxicity of Aflatoxin B1-Mediated Liver and Kidney Derangement in Rats. Sci. Rep. 2022, 12, 7438. [Google Scholar] [CrossRef]
- Watson, M.J.; Berger, P.L.; Banerjee, K.; Frank, S.B.; Tang, L.; Ganguly, S.S.; Hostetter, G.; Winn, M.; Miranti, C.K. Aberrant CREB1 Activation in Prostate Cancer Disrupts Normal Prostate Luminal Cell Differentiation. Oncogene 2021, 40, 3260–3272. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Tsao, C.-M.; Yu, P.-N.; Shih, Y.-L.; Lin, C.-H.; Yan, M.-D. SOX1 Suppresses Cell Growth and Invasion in Cervical Cancer. Gynecol. Oncol. 2013, 131, 174–181. [Google Scholar] [CrossRef]
- Wong, S.H.M.; Fang, C.M.; Chuah, L.-H.; Leong, C.O.; Ngai, S.C. E-Cadherin: Its Dysregulation in Carcinogenesis and Clinical Implications. Crit. Rev. Oncol. Hematol. 2018, 121, 11–22. [Google Scholar] [CrossRef]
- Canel, M.; Serrels, A.; Frame, M.C.; Brunton, V.G. E-Cadherin-Integrin Crosstalk in Cancer Invasion and Metastasis. J. Cell Sci. 2013, 126, 393–401. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th Anniversary of the Open-Access Database of Transcription Factor Binding Profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef]
- Ghouili, F.; Martin, L.J. Cooperative Regulation of Gja1 Expression by Members of the AP-1 Family cJun and cFos in TM3 Leydig and TM4 Sertoli Cells. Gene 2017, 635, 24–32. [Google Scholar] [CrossRef]







| Physicochemical Properties | Lipophilicity | Pharmacokinetics | ||||||
|---|---|---|---|---|---|---|---|---|
| MW (g/mol) | ROTB (n) | HBA (n) | HBD (n) | TPSA (Å) | CLogPo/w | GIA | BBBP | |
| Rule | <500 | ≤10 | <10 | <5 | ≤140 | <5 | - | - |
| P1 | 290.70 | 1 | 4 | 3 | 73.83 | 0.26 | High | No |
| P2 | 452.84 | 4 | 9 | 6 | 152.98 | −1.23 | Low | No |
| P3 | 436.84 | 4 | 8 | 5 | 132.75 | −1.16 | Low | No |
| P5 | 436.84 | 4 | 8 | 5 | 132.75 | −0.84 | Low | No |
| P6 | 450.87 | 5 | 8 | 4 | 121.75 | −0.42 | High | No |
| P7 | 274.70 | 1 | 3 | 2 | 53.60 | 0.91 | High | Yes |
| APN | 290.70 | 1 | 4 | 3 | 73.83 | 0.23 | High | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora, N.; Rosa, M.; Touaibia, M.; Martin, L.J. Effects of Red Sorghum-Derived Deoxyanthocyanidins and Their O-β-D-Glucosides on E-Cadherin Promoter Activity in PC-3 Prostate Cancer Cells. Molecules 2024, 29, 1891. https://doi.org/10.3390/molecules29081891
Mora N, Rosa M, Touaibia M, Martin LJ. Effects of Red Sorghum-Derived Deoxyanthocyanidins and Their O-β-D-Glucosides on E-Cadherin Promoter Activity in PC-3 Prostate Cancer Cells. Molecules. 2024; 29(8):1891. https://doi.org/10.3390/molecules29081891
Chicago/Turabian StyleMora, Nathalie, Maxence Rosa, Mohamed Touaibia, and Luc J. Martin. 2024. "Effects of Red Sorghum-Derived Deoxyanthocyanidins and Their O-β-D-Glucosides on E-Cadherin Promoter Activity in PC-3 Prostate Cancer Cells" Molecules 29, no. 8: 1891. https://doi.org/10.3390/molecules29081891
APA StyleMora, N., Rosa, M., Touaibia, M., & Martin, L. J. (2024). Effects of Red Sorghum-Derived Deoxyanthocyanidins and Their O-β-D-Glucosides on E-Cadherin Promoter Activity in PC-3 Prostate Cancer Cells. Molecules, 29(8), 1891. https://doi.org/10.3390/molecules29081891