The Potential Antinociceptive Effect and Mechanism of Cannabis sativa L. Extract on Paclitaxel-Induced Neuropathic Pain in Rats Uncovered by Multi-Omics Analysis
Abstract
:1. Introduction
2. Results
2.1. Chemical Composition of Hemp Extract (JG)
2.2. Antinociceptive Effect of Hemp Extract (JG) on PTX-Induced Neuropathic Rats
2.2.1. Effect of Hemp Extract (JG) on Neurobehavioral Changes of PTX-Induced Neuropathic Rats
2.2.2. Effect of Hemp Extract (JG) on Inflammatory Cytokines of PTX-Induced Neuropathic Rats
2.3. Transcriptome Analysis
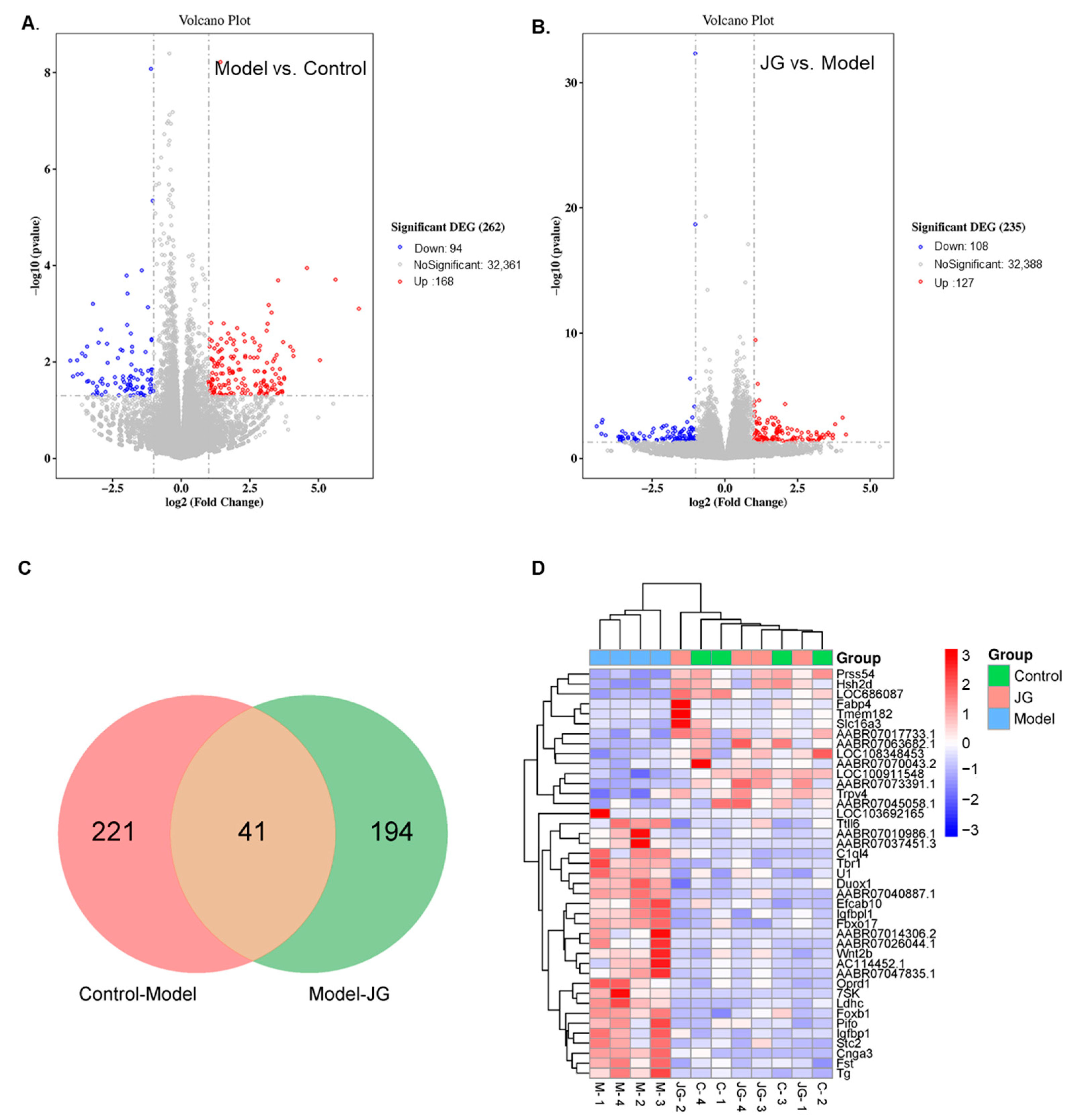
2.3.1. Identification of Differentially Expressed Genes (DEGs)
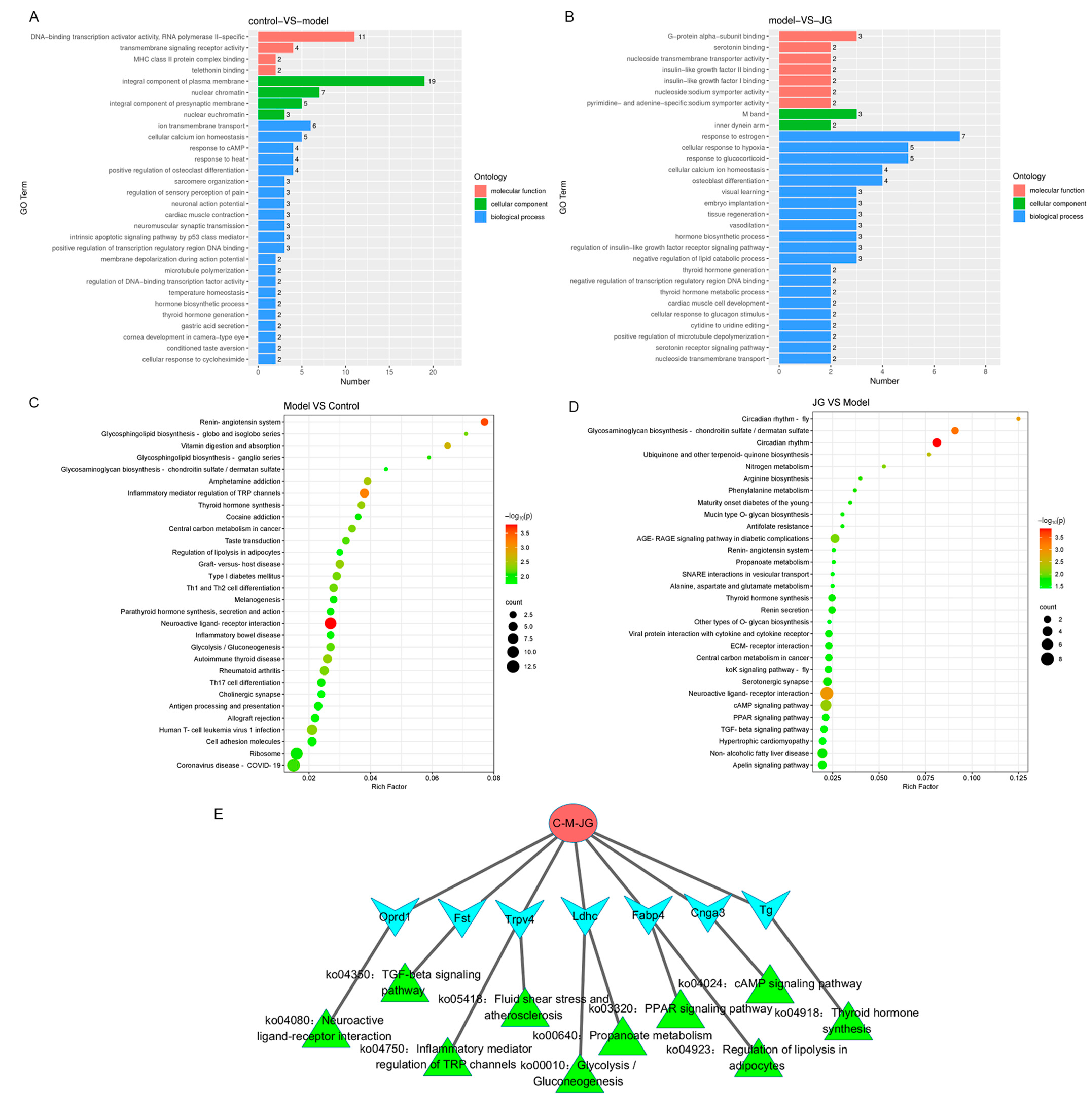
2.3.2. Gene Ontology (GO) Enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of DEGs
2.4. Untargeted Metabolome Analysis
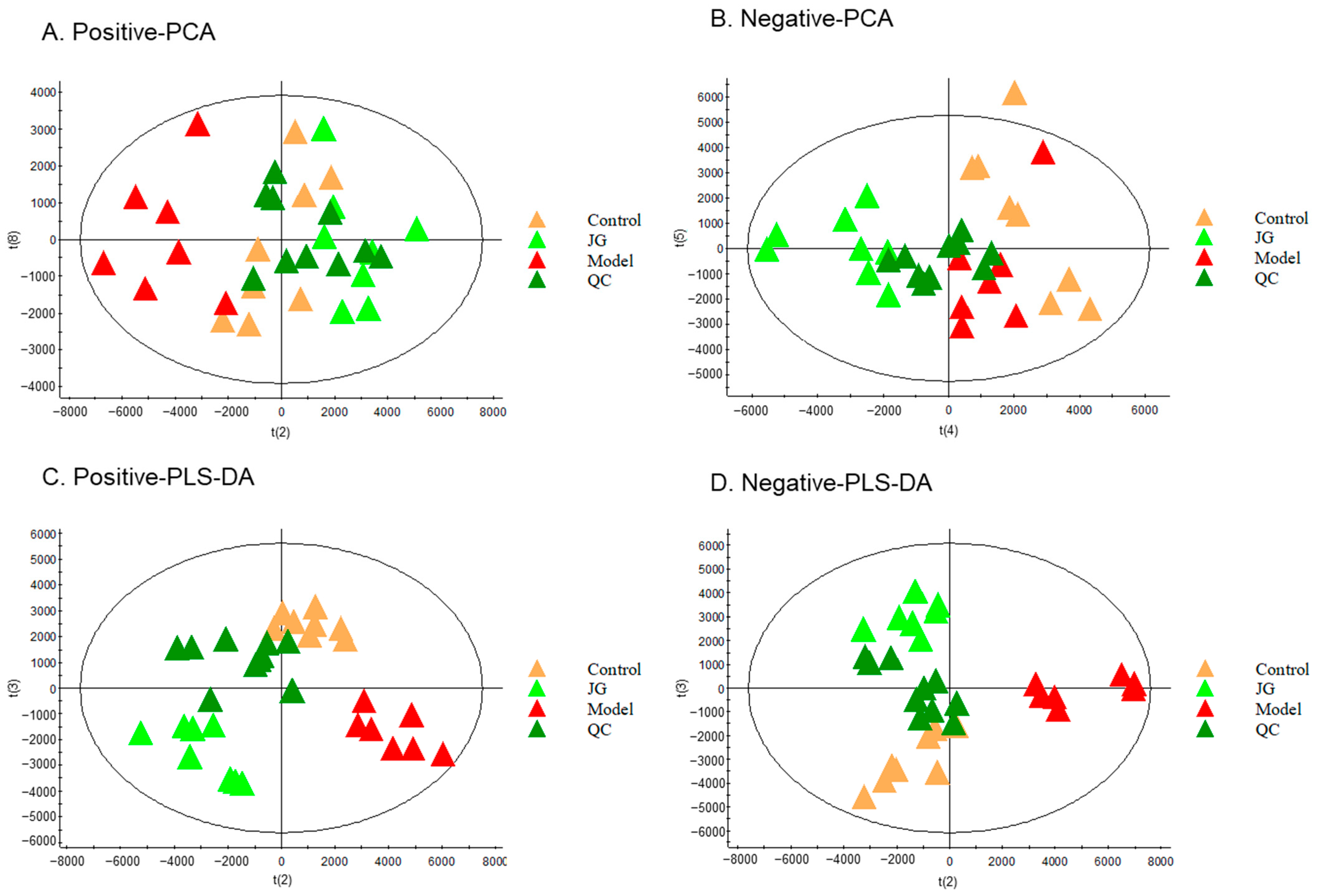
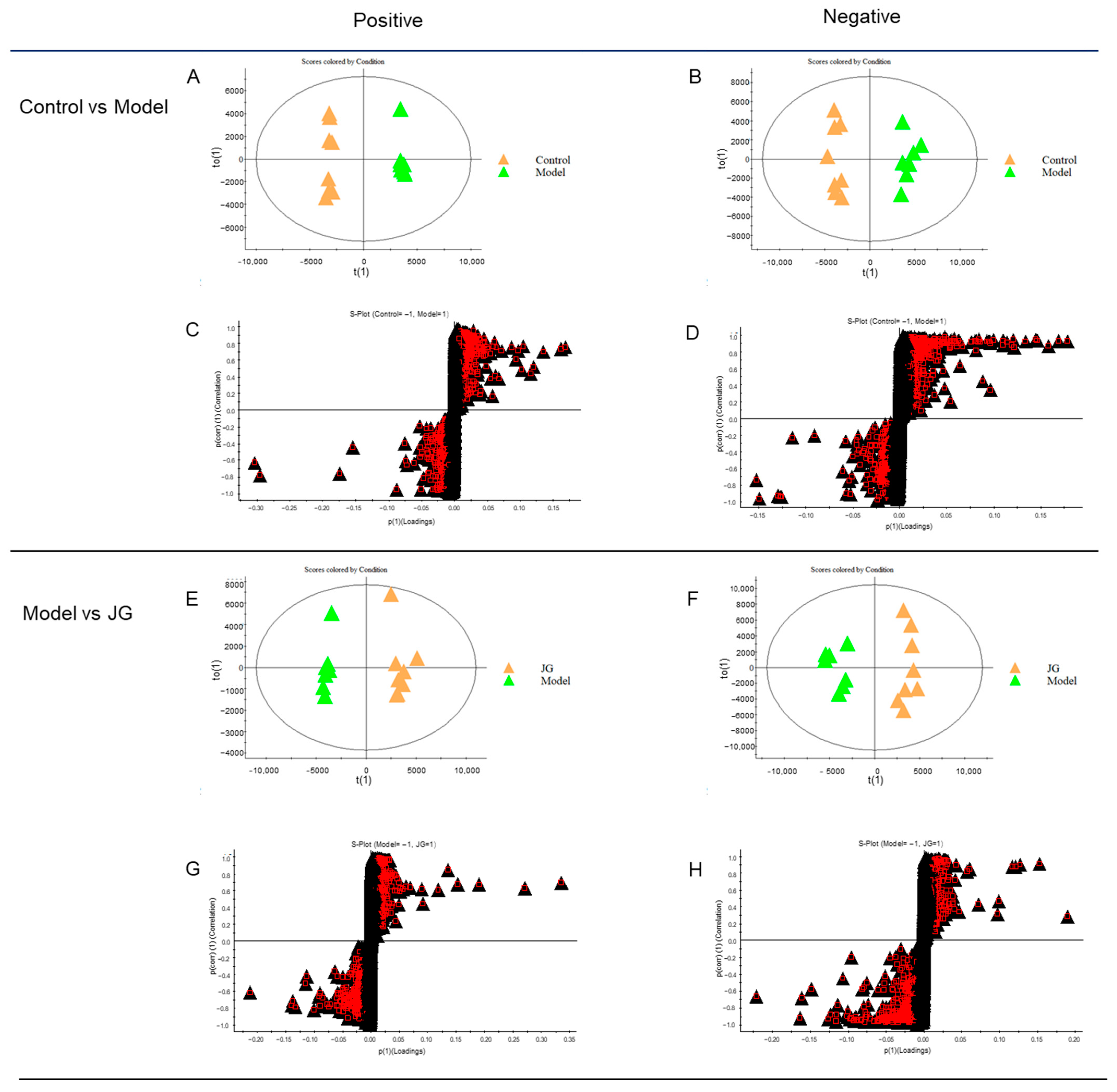
2.4.1. Identification of Differential Metabolites (DMs)
2.4.2. Metabolic Pathway Enrichment Analysis
2.5. Gut Microbiota Analysis
2.5.1. Gut Microbiota Diversity Variation
2.5.2. Analysis of Species Abundance
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Preparation and Analysis of Cannabis sativa L. Extract (JG)
4.3. Animals and Treatment
4.4. Behavioral Tests
4.4.1. Mechanical Allodynia
4.4.2. Thermal Hyperalgesia
4.5. Enzyme Linked Immunosorbent Assay
4.6. Transcriptomic Analysis of Spinal Cord
4.6.1. RNA Extraction and Sequencing
4.6.2. Data Processing and Bioinformatics Analysis
4.7. Untargeted Metabolomic Analysis of Serum Samples
4.7.1. Sample Preparation and UPLC-Q-TOF/MS Metabolomics Analysis
4.7.2. Data Processing and Multivariate Analysis
4.8. Gut Microbiota Analysis of Cecum Content
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hourfane, S.; Mechqoq, H.; Bekkali, A.Y.; Rocha, J.M.; El Aouad, N. A Comprehensive Review on Cannabis sativa Ethnobotany, Phytochemistry, Molecular Docking and Biological Activities. Plants 2023, 12, 1245. [Google Scholar] [CrossRef]
- Radwan, M.M.; Chandra, S.; Gul, S.; ElSohly, M.A. Cannabinoids, Phenolics, Terpenes and Alkaloids of Cannabis. Molecules 2021, 26, 2774. [Google Scholar] [CrossRef]
- Shehata, I.; Hashim, A.; Elsaeidy, A.; Nair, A.; Urits, I.; Viswanath, O.; Kaye, A.; Habib, M. Cannabinoids and Their Role in Chronic Pain Treatment: Current Concepts and a Comprehensive Review. Health Psychol. Res. 2022, 10, 35848. [Google Scholar] [CrossRef] [PubMed]
- Bakogeorgos, M.; Georgoulias, V. Risk-reduction and treatment of chemotherapy-induced peripheral neuropathy. Expert Rev. Anticancer Ther. 2017, 17, 1045–1060. [Google Scholar] [CrossRef]
- Addington, J.; Freimer, M. Chemotherapy-induced peripheral neuropathy: An update on the current understanding. F1000 Res. 2016, 5, 1466. [Google Scholar] [CrossRef]
- Cavaletti, G.; Marmiroli, P. Chemotherapy-induced peripheral neurotoxicity. Nat. Rev. Neurol. 2010, 6, 657–666. [Google Scholar] [CrossRef]
- Gornstein, E.; Schwarz, T.L. The paradox of paclitaxel neurotoxicity: Mechanisms and unanswered questions. Neuropharmacology 2014, 76, 175–183. [Google Scholar] [CrossRef]
- Wu, Z.; Toro, G.; Xu, G.; Dang, D.; Prater, C.; Yang, Q. Paclitaxel Inhibits KCNQ Channels in Primary Sensory Neurons to Initiate the Development of Painful Peripheral Neuropathy. Cells 2022, 11, 4067. [Google Scholar] [CrossRef]
- Cella, D.; Peterman, A.; Hudgens, S.; Webster, K.; Socinski, M.A. Measuring the side effects of taxane therapy in oncology: The functional assessment of cancer therapy-taxane (FACT-taxane). Cancer 2003, 98, 822–831. [Google Scholar] [CrossRef]
- Altiparmak, B.; Cil, H.; Celebi, N. Efeito da melatonina sobre o efeito colateral de sonolência diurna da gabapentina em pacientes adultos com dor neuropática [Effect of melatonin on the daytime sleepiness side-effect of gabapentin in adults patients with neuropathic pain]. Braz. J. Anesthesiol. 2019, 69, 137–143. [Google Scholar] [CrossRef]
- Campos, R.M.P.; Aguiar, A.F.L.; Paes-Colli, Y.; Trindade, P.M.P.; Ferreira, B.K.; de Melo Reis, R.A.; Sampaio, L.S. Cannabinoid Therapeutics in Chronic Neuropathic Pain: From Animal Research to Human Treatment. Front. Physiol. 2021, 12, 785176. [Google Scholar] [CrossRef] [PubMed]
- Maayah, Z.H.; Takahara, S.; Ferdaoussi, M.; Dyck, J.R.B. The molecular mechanisms that underpin the biological benefits of full-spectrum cannabis extract in the treatment of neuropathic pain and inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165771. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.J.; Ramirez, M.D.; Neelakantan, H.; Walker, E.A. Cannabidiol prevents the development of cold and mechanical allodynia in paclitaxel-treated female C57Bl6 mice. Anesth. Analg. 2011, 113, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.J.; McAllister, S.D.; Kawamura, R.; Murase, R.; Neelakantan, H.; Walker, E.A. Cannabidiol inhibits paclitaxel-induced neuropathic pain through 5-HT(1A) receptors without diminishing nervous system function or chemotherapy efficacy. Br. J. Pharmacol. 2014, 171, 636–645. [Google Scholar] [CrossRef]
- Alkislar, I.; Miller, A.R.; Hohmann, A.G.; Sadaka, A.H.; Cai, X.; Kulkarni, P.; Ferris, C.F. Inhaled Cannabis Suppresses Chemotherapy-Induced Neuropathic Nociception by Decoupling the Raphe Nucleus: A Functional Imaging Study in Rats. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2021, 6, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Kumar Kalvala, A.; Bagde, A.; Arthur, P.; Kumar Surapaneni, S.; Ramesh, N.; Nathani, A.; Singh, M. Role of Cannabidiol and Tetrahydrocannabivarin on Paclitaxel-induced neuropathic pain in rodents. Int. Immunopharmacol. 2022, 107, 108693. [Google Scholar] [CrossRef]
- King, K.M.; Myers, A.M.; Soroka-Monzo, A.J.; Tuma, R.F.; Tallarida, R.J.; Walker, E.A.; Ward, S.J. Single and combined effects of Δ9 -tetrahydrocannabinol and cannabidiol in a mouse model of chemotherapy-induced neuropathic pain. Br. J. Pharmacol. 2017, 174, 2832–2841. [Google Scholar] [CrossRef] [PubMed]
- Comelli, F.; Giagnoni, G.; Bettoni, I.; Colleoni, M.; Costa, B. Antihyperalgesic effect of a Cannabis sativa extract in a rat model of neuropathic pain: Mechanisms involved. Phytother. Res. 2008, 22, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Comelli, F.; Bettoni, I.; Colleoni, M.; Giagnoni, G.; Costa, B. Beneficial effects of a Cannabis sativa extract treatment on diabetes-induced neuropathy and oxidative stress. Phytother. Res. 2009, 23, 1678–1684. [Google Scholar] [CrossRef]
- Pattnaik, F.; Hans, N.; Patra, B.R.; Nanda, S.; Kumar, V.; Naik, S.N.; Dalai, A.K. Valorization of Wild-Type Cannabis indica by Supercritical CO2 Extraction and Insights into the Utilization of Raffinate Biomass. Molecules 2022, 28, 207. [Google Scholar] [CrossRef]
- Gallo-Molina, A.C.; Castro-Vargas, H.I.; Garzón-Méndez, W.F.; Ramírez, J.A.; Monroy, Z.J.; King, J.W.; Parada-Alfonso, F. Extraction, isolation and purification of tetrahydrocannabinol from the Cannabis sativa L. plant using supercritical fluid extraction and solid phase extraction. J. Supercrit. Fluids 2019, 146, 208–216. [Google Scholar] [CrossRef]
- Zhang, H.W.; Lv, C.; Zhang, L.J.; Guo, X.; Shen, Y.W.; Nagle, D.G.; Zhou, Y.D.; Liu, S.H.; Zhang, W.D.; Luan, X. Application of omics- and multi-omics-based techniques for natural product target discovery. Biomed. Pharmacother. 2021, 141, 111833. [Google Scholar]
- Sun, J.; Zhou, J.; Luo, J.J.; Xu, Y.H.; Hua, M.L. Determination of Chemical Constituents in Flowers and Leaves of Hemp by GC-MS and UPLC-Q-TOF-MS. Chin. J. Pharm. 2022, 10, 1465–1481. [Google Scholar]
- Ferrer, I. Chapter Fourteen-Analyses of cannabinoids in hemp oils by LC/Q-TOF-MS. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 90, pp. 415–452. [Google Scholar] [CrossRef]
- McLeary, F.; Davis, A.; Rudrawar, S.; Perkins, A.; Anoopkumar-Dukie, S. Mechanisms underlying select chemotherapeutic-agent-induced neuroinflammation and subsequent neurodegeneration. Eur. J. Pharmacol. 2019, 842, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, Y.; Ye, X.; Zhang, N.; Pan, L.; Wang, B. TRP (transient receptor potential) ion channel family: Structures, biological functions and therapeutic interventions for diseases. Signal Transduct. Target. Ther. 2023, 8, 261. [Google Scholar] [CrossRef]
- Singh, R.; Adhya, P.; Sharma, S.S. Redox-sensitive TRP channels: A promising pharmacological target in chemotherapy-induced peripheral neuropathy. Expert Opin. Ther. Targets 2021, 25, 529–545. [Google Scholar]
- Sun, W.; Yang, S.; Wu, S.; Ba, X.; Xiong, D.; Xiao, L.; Hao, Y. Transcriptome analysis reveals dysregulation of inflammatory and neuronal function in dorsal root ganglion of paclitaxel-induced peripheral neuropathy rats. Mol. Pain 2023, 19, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Yin, C.; Liu, B.; Nie, H.; Wang, J.; Zeng, D.; Chen, R.; He, X.; Fang, J.; Du, J.; et al. Transcriptome profiling of long noncoding RNAs and mRNAs in spinal cord of a rat model of paclitaxel-induced peripheral neuropathy identifies potential mechanisms mediating neuroinflammation and pain. J. Neuroinflamm. 2021, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Lafaye, G.; Desterke, C.; Marulaz, L.; Benyamina, A. Cannabidiol affects circadian clock core complex and its regulation in microglia cells. Addict. Biol. 2019, 24, 921–934. [Google Scholar] [CrossRef]
- Edwards, D.; Filbey, F.M. Are Sweet Dreams Made of These? Understanding the Relationship Between Sleep and Cannabis Use. Cannabis Cannabinoid Res. 2021, 6, 462–473. [Google Scholar] [CrossRef]
- Echeverry, S.; Shi, X.Q.; Haw, A.; Liu, H.; Zhang, Z.W.; Zhang, J. Transforming growth factor-beta1 impairs neuropathic pain through pleiotropic effects. Mol. Pain 2009, 5, 1744–8069. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.; Ruviaro, N.A.; Trevisan, G. TRPV4 Role in Neuropathic Pain Mechanisms in Rodents. Antioxidants 2022, 12, 24. [Google Scholar] [CrossRef]
- Muller, C.; Morales, P.; Reggio, P.H. Cannabinoid Ligands Targeting TRP Channels. Front. Mol. Neurosci. 2019, 11, 487. [Google Scholar] [CrossRef]
- Silva-Cardoso, G.K.; Lazarini-Lopes, W.; Hallak, J.E.; Crippa, J.A.; Zuardi, A.W.; Garcia-Cairasco, N.; Leite-Panissi, C.R.A. Cannabidiol effectively reverses mechanical and thermal allodynia, hyperalgesia, and anxious behaviors in a neuropathic pain model: Possible role of CB1 and TRPV1 receptors. Neuropharmacology 2021, 197, 108712. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Z.; Xu, W.J.; Deng, B.; Liu, S.D.; Deng, C.; Wu, M.Y.; Gao, Y.; Jia, L.Q. Wen-Luo-Tong Decoction Attenuates Paclitaxel-Induced Peripheral Neuropathy by Regulating Linoleic Acid and Glycerophospholipid Metabolism Pathways. Front. Pharmacol. 2018, 9, 956. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Wang, Y.; Zhang, P.; Yuan, Y.; Zhang, Y.; Chen, G. Gut microbiota regulates neuropathic pain: Potential mechanisms and therapeutic strategy. J. Headache Pain 2020, 21, 103. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Zhou, Z.; Liang, Y.; Cheng, X.; Li, Y.; Teng, W.; Zhao, M.; Liu, C.; Guan, M.; Zhao, C. Targeting strategies for chemotherapy-induced peripheral neuropathy: Does gut microbiota play a role? Crit. Rev. Microbiol. 2019, 45, 369–393. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chen, M.; Zheng, C.; Yinglin, B.; Zhu, B. Fecal Microbiota Transplantation Alleviated Paclitaxel-Induced Peripheral Neuropathy by Interfering with Astrocytes and TLR4/p38MAPK Pathway in Rats. J. Pain Res. 2023, 16, 2419–2432. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, C.; Corleto, J.; Ruegger, P.M.; Logan, G.D.; Peacock, B.B.; Mendonca, S.; Yamaki, S.; Adamson, T.; Ermel, R.; McKemy, D.; et al. Dominant Role of the Gut Microbiota in Chemotherapy Induced Neuropathic Pain. Sci. Rep. 2019, 9, 20324. [Google Scholar] [CrossRef]
- Loman, B.R.; Jordan, K.R.; Haynes, B.; Bailey, M.T.; Pyter, L.M. Chemotherapy-induced neuroinflammation is associated with disrupted colonic and bacterial homeostasis in female mice. Sci. Rep. 2019, 9, 16490. [Google Scholar] [CrossRef]
- Su, T.; Yin, X.; Ren, J.; Lang, Y.; Zhang, W.; Cui, L. Causal relationship between gut microbiota and myasthenia gravis: A bidirectional mendelian randomization study. Cell Biosci. 2023, 13, 204. [Google Scholar] [CrossRef] [PubMed]
- Cirstea, M.; Radisavljevic, N.; Finlay, B.B. Good Bug, Bad Bug: Breaking through Microbial Stereotypes. Cell Host Microbe 2018, 23, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Abdelbary, M.M.H.; Hatting, M.; Bott, A.; Dahlhausen, A.; Keller, D.; Trautwein, C.; Conrads, G. The oral-gut axis: Salivary and fecal microbiome dysbiosis in patients with inflammatory bowel disease. Front. Cell Infect. Microbiol. 2022, 12, 1010853. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.J.; Jia, Y.X.; Yue, W.; Li, X.L.; Ni, B.B. Post-decarboxylation and supercritical fluid extraction of cannabidiol from hemp. Contemp. Chem. Ind. 2021, 50, 2596–2599. [Google Scholar]
- Lewis, M.M.; Yang, Y.; Wasilewski, E.; Clarke, H.A.; Kotra, L.P. Chemical Profiling of Medical Cannabis Extracts. ACS Omega 2017, 2, 6091–6103. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.H.; Zhou, J.; Xu, Y.H.; Xiao, F.K.; Hua, M.L. Determination of Tetrahydrocannabinol, Cannabidiol and Cannabidiolic Acid in Domestic Hemp Leaves and Flowers by HPLC. Chin. J. Pharm. 2022, 53, 95–100. [Google Scholar]
- Polomano, R.C.; Mannes, A.J.; Clark, U.S.; Bennett, G.J. A painful peripheral neuropathy in the rat produced by the chemotherapeutic drug, paclitaxel. Pain 2001, 94, 293–304. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar]
- Liao, J.; Zhang, Y.; Zhang, W.; Zeng, Y.; Zhao, J.; Zhang, J.; Yao, T.; Li, H.; Shen, X.; Wu, G.; et al. Different software processing affects the peak picking and metabolic pathway recognition of metabolomics data. J. Chromatogr. A 2023, 1687, 463700. [Google Scholar] [CrossRef]







| No. | RT (min) | Tentative Identification | Formula | m/z | Molecular Ion Peak | Adducts (Positive) | Adducts (Negative) |
|---|---|---|---|---|---|---|---|
| 1 | 8.10 | Cannabispiradienone [2] | C15H14O3 | 242 | 243.1405 [M + H]+, 241.9238 [M − H]− | 227.1516 | |
| 2 | 8.61 | Cannabispireone [2,23] | C15H16O3 | 244 | 245.1581 [M + H]+, 243.1261 [M − H]− | 229.1683 | |
| 3 | 8.91 | Cannabispiran [2,23] | C15H18O3 | 246 | 247.1710 [M + H]+, 245.1418, [M − H]− | 189.1302 | 161.0751 |
| 4 | 9.97 | CBE-C3 [23] | C19H26O3 | 302 | 303.1593 [M + H]+, 301.1361 [M − H]− | 271.1359, 257.1559, 243.1405 | 286.1120, 271.0879, 243.9218 |
| 5 | 10.15 | Cannabichromanone-D [2] | C21H28O3 | 328 | 329.2095 [M + H]+, 373.2010 [M + 45]−, 327.1910 [M − H]− | 301.1425, 289.2181, 273.1477 | 293.2042, 236.1302 |
| 6 | 10.48 | CBT-C3 [2] | C19H26O4 | 318 | 319.2248 [M + H]+, 317.2080 [M − H]− | 301.2136, 285.1125 | 299.1954, 281.1802, 257.1778, 163.0930 |
| 7 | 10.65 | CBGAM [23] | C23H34O4 | 374 | 749.4123 [2M + Na]+, 397.2253 [M + Na]+, 375.2461 [M + H]+, 373.2367 [M − H]− | 357.2377, 339.2273, 275.1671 | 329.2462, 311.2404 |
| 8 | 11.18 | CBT-C5 [23] | C21H30O4 | 346 | 347.2566 [M + H]+, 345.2388 [M − H]− | 329.2466, 311.2323, 279.2690, 231.1763 | 327.2280, 313.2704, 289.1752, 271.1621 |
| 9 | 11.36 | CBR [23] | C21H32O4 | 348 | 371.2651 [M + Na]+, 349.2691 [M + H]+, 347.2583 [M − H]− | 327.1929, 313.499, 297.2774, 279.2690 | 329.2462, 311.2513, 295.2238 |
| 10 | 11.50 | CBCV [23,24] | C19H26O2 | 286 | 287.2380 [M + H]+, 285.2121 [M − H]− | 201.2038 | 191.1242 |
| 11 | 11.79 | Cannflavin B [2] | C21H20O6 | 368 | 369.1666 [M + H]+, 367.1573 [M − H]− | 313.1050, 291.2330, 193.1616 | 331.2632 |
| 12 | 12.08 | CBEAB-C3 [2] | C20H26O5 | 346 | 369.1666 [M + Na]+, 347.1804 [M + H]+, 345.2083 [M − H]− | 313.2499, 201.2136, 715.2702 | 311.2332, 329.2462, 191.1242 |
| 13 | 12.69 | CBT-OEt-C3 [2] | C21H30O4 | 346 | 347.2566 [M + H]+, 345.2388 [M − H]− | 329.2429, 277.2551 | 309.2170, 285.2121, 279.1648, 191.1242 |
| 14 | 13.25 | CBDVA [23,24] | C20H26O4 | 330 | 331.2621 [M + H]+, 329.2425 [M − H]− | 313.2499, 275.2383 | 325.2123, 309.2350 |
| 15 | 13.83 | 4-Acetoxy-CBC [2] | C23H32O4 | 372 | 373.2312 [M + H]+, 371.2218 [M − H]− | 355.2217, 331.2323, 313.2173, 279.2690 | 329.2091, 311.1971, 295.2555, 277.2453 |
| 16 | 14.09 | Epoxycannabigerol [2] | C21H32O3 | 332 | 333.2763 [M + H]+, 331.2595 [M − H]− | 315.2668, 279.2690 | 325.2123, 295.2590, 313.2487 |
| 17 | 14.54 | CBDV [23,24] | C19H26O2 | 286 | 287.2380 [M + H]+, 285.2121 [M − H]− | 231.1763 | 217.1443 |
| 18 | 14.66 | Cannabimovone [2] | C21H30O4 | 346 | 347.2566 [M + H]+, 345.2388 [M − H]− | 329.2466 | 293.2393 |
| 19 | 15.60 | CBTT [2] | C21H30O5 | 362 | 363.2459 [M + H]+, 361.2252 [M − H]− | 345.2386, 327.2299, | |
| 20 | 16.10 | CBGM [23] | C22H34O2 | 330 | 331.2621 [M + H]+, 329.2425 [M − H]− | 313.2499 | 311.2079, 243.9218 |
| 21 | 16.79 | CBND-C3 [23] | C19H22O2 | 282 | 283.205 [M + H]+, 281.1802 [M − H]− | 271.2066 | 243.9218, 311.2007 |
| 22 | 17.07 | CBDA [23,24] | C22H30O4 | 358 | 341.2466 [M + H − H2O]+, 357.2701 [M − H]− | 329.2466, 283.2085 | 311.2043, 281.1802 |
| 23 | 17.20 | Canflavin C [2] | C26H28O6 | 436 | 437.2217 [M + H]+, 435.2238 [M − H]− | 383.2517, 359.2556, 313.1050 | 399.2595, 327.2280, 295.2590, 243.9218 |
| 24 | 18.15 | CBG [23,24] | C21H32O2 | 316 | 317.2828 [M + H]+, 315.2634 [M − H]− | 287.238 | |
| 25 | 18.63 | CBD [23,24] | C21H30O2 | 314 | 315.2668 [M + H]+, 313.2451 [M − H]− | 259.2076 | 313.2270, 245.1771 |
| 26 | 19.45 | CBCN-C5 [2] | C20H28O4 | 332 | 333.2763 [M + H]+, 331.2595 [M − H]− | 315.2668, 259.2076 | 325.2160, 293.2077, 243.9218 |
| 27 | 22.78 | CBN [23,24] | C21H26O2 | 310 | 311.2359 [M + H]+, 309.2134 [M − H]− | 293.2269 | 277.2453 |
| 28 | 24.63 | CBL [23,24] | C21H30O2 | 314 | 315.2668 [M + H]+ | ||
| 29 | 25.37 | CBDM [2] | C22H32O2 | 328 | 329.2466 [M + H]+, 327.2280 [M − H]− | 315.2668 | 273.1755, 243.9218 |
| 30 | 26.02 | CBC [23,24] | C21H30O2 | 314 | 315.2688 [M + H]+ | ||
| 31 | 27.57 | Δ8-THC [23] | C21H30O2 | 314 | 315.2668 [M + H]+ | 279.2572, 243.2218 | |
| 32 | 28.52 | Δ9-THC [23] | C21H30O2 | 314 | 315.2668 [M + H]+, 313.2487 [M − H]− | 259.2076 | 205.1422, 243.9218 |
| 33 | 32.89 | CBT-OEt-C5 [23] | C23H34O4 | 374 | 397.4129 [M + Na]+, 373.2762 [M − H]− | 357.2416, 331.2994, 207.1803 | 329.2834 |
| Phytocannabinoid | Abbreviation | Content |
|---|---|---|
| Cannabidiol | CBD | 30.10% 1 |
| Cannabidiolic acid | CBDA | 0.90% 2 |
| Cannabidivarin | CBDV | 5.74% 2 |
| Δ9-tetrahydrocannabinol | Δ9-THC | 2.42% 2 |
| Number | JG Regulation | Gene Symbol |
|---|---|---|
| 41 | Down (27) | Ttll6, Wnt2b, Stc2, AABR07010986.1, Cnga3, Igfbp1, U1, Tbr1, Tg, Oprd1, Foxb1, Efcab10, Igfbpl1, Fst, Ldhc, Pifo, Fbxo17, AABR07047835.1, Duox1, AABR07040887.1, 7SK, AC114452.1, AABR07014306.2, LOC103692165, C1ql4, AABR07026044.1, AABR07037451.3 |
| Up (14) | Trpv4, Fabp4, Prss54, LOC686087, AABR07073391.1, LOC108348453, LOC100911548, Hsh2d, Tmem182, AABR07045058.1, Slc16a3, AABR07017733.1, AABR07070043.2, AABR07063682.1 |
| No. | Rt (min) | ID | Formula | m/z | Description |
|---|---|---|---|---|---|
| 1 | 7.88 | HMDB39412 | C54H88O23 | 1127.55 | Araliasaponin V |
| 2 | 8.04 | HMDB60075 | C15H13I2NO7S | 1254.69 | 3′,5′-Diiodo-l-thyronine 4′-O-sulfate |
| 3 | 8.04 | HMDB35746 | C57H92O26 | 1215.58 | Hovenoside D |
| 4 | 8.12 | HMDB04936 | C61H109N3O26 | 1298.71 | Ganglioside GM2 (d18:1/12:0) |
| 5 | 8.12 | HMDB32780 | C59H96O27 | 1259.60 | Jujuboside C |
| 6 | 8.85 | HMDB29513 | C22H16O15S | 575.01 | Myricatin |
| 7 | 14.40 | HMDB11484 | C25H46NO7P | 502.29 | LysoPE(0:0/20:3(11Z,14Z,17Z)) |
| 8 | 14.40 | HMDB10387 | C26H48NO7P | 562.31 | LysoPC(18:3(6Z,9Z,12Z)) |
| 9 | 14.40 | HMDB30459 | C29H40N8O5 | 579.31 | Hordatine B |
| 10 | 14.49 | HMDB10397 | C28H48NO7P | 586.32 | LysoPC(20:5(5Z,8Z,11Z,14Z,17Z)) |
| 11 | 14.49 | HMDB35388 | C34H48O7 | 603.31 | (24E)-3α,15α-Diacetoxy-23-oxo-7,9(11),24-lanostatrien-26-oic acid |
| 12 | 15.11 | HMDB11129 | C23H48NO7P | 526.32 | LysoPE(0:0/18:0) |
| 13 | 15.14 | HMDB11914 | C55H102N2O21 | 1107.68 | Ganglioside GM3 (d18:0/14:0) |
| 14 | 15.14 | HMDB35292 | C34H50O7 | 605.32 | Ganodermic acid P2 |
| 15 | 15.36 | HMDB31906 | C37H66O2 | 1084.01 | Muridienin 4 |
| 16 | 15.36 | HMDB38189 | C31H44O6 | 1023.63 | Carindone |
| 17 | 15.42 | HMDB30317 | C28H37NO9 | 1107.49 | Petasinoside |
| 18 | 15.42 | HMDB38337 | C38H66O2 | 1108.01 | Helianyl octanoate |
| 19 | 15.42 | HMDB41384 | C23H20O13 | 522.13 | Luteolin 3′-(4″-acetylglucuronide) |
| 20 | 15.45 | HMDB35310 | C30H44O8 | 1087.59 | Ganoderic acid I |
| 21 | 15.45 | HMDB35356 | C56H88O20 | 1063.59 | Pithecelloside |
| 22 | 15.73 | HMDB02513 | C30H48O9 | 1103.66 | Lithocholate 3-O-glucuronide |
| 23 | 15.73 | HMDB10386 | C26H50NO7P | 1037.67 | LysoPC(18:2(9z,12z)) |
| 24 | 15.76 | HMDB10356 | C24H32O12S | 543.16 | Estriol 3-sulfate 16-glucuronide |
| 25 | 16.04 | HMDB29766 | C24H40O11 | 522.29 | (3S,7E,9R)-4,7-Megastigmadiene-3,9-diol 9-[apiosyl-(1->6)-glucoside] |
| 26 | 16.04 | HMDB34731 | C27H45NO2 | 454.31 | Tomatidine |
| 27 | 16.14 | HMDB01980 | C46H65N13O12S2 | 1036.41 | Vasopressin |
| 28 | 16.14 | HMDB10382 | C24H50NO7P | 1035.67 | LysoPC(16:0) |
| 29 | 16.19 | HMDB10364 | C27H38O9 | 524.30 | 11-Hydroxyprogesterone 11-glucuronide |
| 30 | 16.23 | HMDB41366 | C29H44O8 | 1041.61 | 24,25-Diacetylvulgaroside |
| 31 | 16.23 | HMDB29341 | C27H40N4O4 | 991.61 | Ceanothine D |
| 32 | 16.50 | HMDB40684 | C37H47NO4 | 614.34 | Janthitrem C |
| 33 | 16.76 | HMDB40914 | C56H85N15O12 | 1140.64 | Mytilus small cardioactive peptide |
| 34 | 16.88 | HMDB30199 | C28H44N4O4 | 1045.68 | Frangulanine |
| 35 | 17.69 | HMDB13622 | C19H36O2 | 341.27 | Nonadeca-10(Z)-enoic acid |
| 36 | 18.24 | HMDB11491 | C27H54NO7P | 536.37 | LysoPE(0:0/22:1(13Z)) |
| 37 | 18.47 | HMDB33428 | C16H18N2O | 507.28 | (+)-Setoclavine |
| 38 | 19.30 | HMDB29328 | C25H29NO10 | 526.17 | Gravacridonediol glucoside |
| 39 | 19.30 | HMDB07925 | C44H74NO8P | 814.48 | PC(14:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Yao, L.; Guo, Y.; Shi, C.; Zhou, J.; Hua, M. The Potential Antinociceptive Effect and Mechanism of Cannabis sativa L. Extract on Paclitaxel-Induced Neuropathic Pain in Rats Uncovered by Multi-Omics Analysis. Molecules 2024, 29, 1958. https://doi.org/10.3390/molecules29091958
Xu Y, Yao L, Guo Y, Shi C, Zhou J, Hua M. The Potential Antinociceptive Effect and Mechanism of Cannabis sativa L. Extract on Paclitaxel-Induced Neuropathic Pain in Rats Uncovered by Multi-Omics Analysis. Molecules. 2024; 29(9):1958. https://doi.org/10.3390/molecules29091958
Chicago/Turabian StyleXu, Yunhui, Lijuan Yao, Yuhan Guo, Chenfeng Shi, Jing Zhou, and Moli Hua. 2024. "The Potential Antinociceptive Effect and Mechanism of Cannabis sativa L. Extract on Paclitaxel-Induced Neuropathic Pain in Rats Uncovered by Multi-Omics Analysis" Molecules 29, no. 9: 1958. https://doi.org/10.3390/molecules29091958
APA StyleXu, Y., Yao, L., Guo, Y., Shi, C., Zhou, J., & Hua, M. (2024). The Potential Antinociceptive Effect and Mechanism of Cannabis sativa L. Extract on Paclitaxel-Induced Neuropathic Pain in Rats Uncovered by Multi-Omics Analysis. Molecules, 29(9), 1958. https://doi.org/10.3390/molecules29091958






