Ultrasonic-Assisted Extraction of Xanthorrhizol from Curcuma xanthorrhiza Roxb. Rhizomes by Natural Deep Eutectic Solvents: Optimization, Antioxidant Activity, and Toxicity Profiles
Abstract
:1. Introduction
2. Results and Discussion
2.1. NADES Preparation
2.2. Evaluation of NADES Antioxidative Activity
2.3. Evaluation of NADES Toxicity
2.4. NADES Extraction of Xanthorrhizol and Determination of the Optimal Conditions Using an Ultrasound-Assisted Extraction Process
2.5. Xanthorrhizol Extraction by GluLA and Comparison with Ethanol
2.5.1. Ethanol Maceration
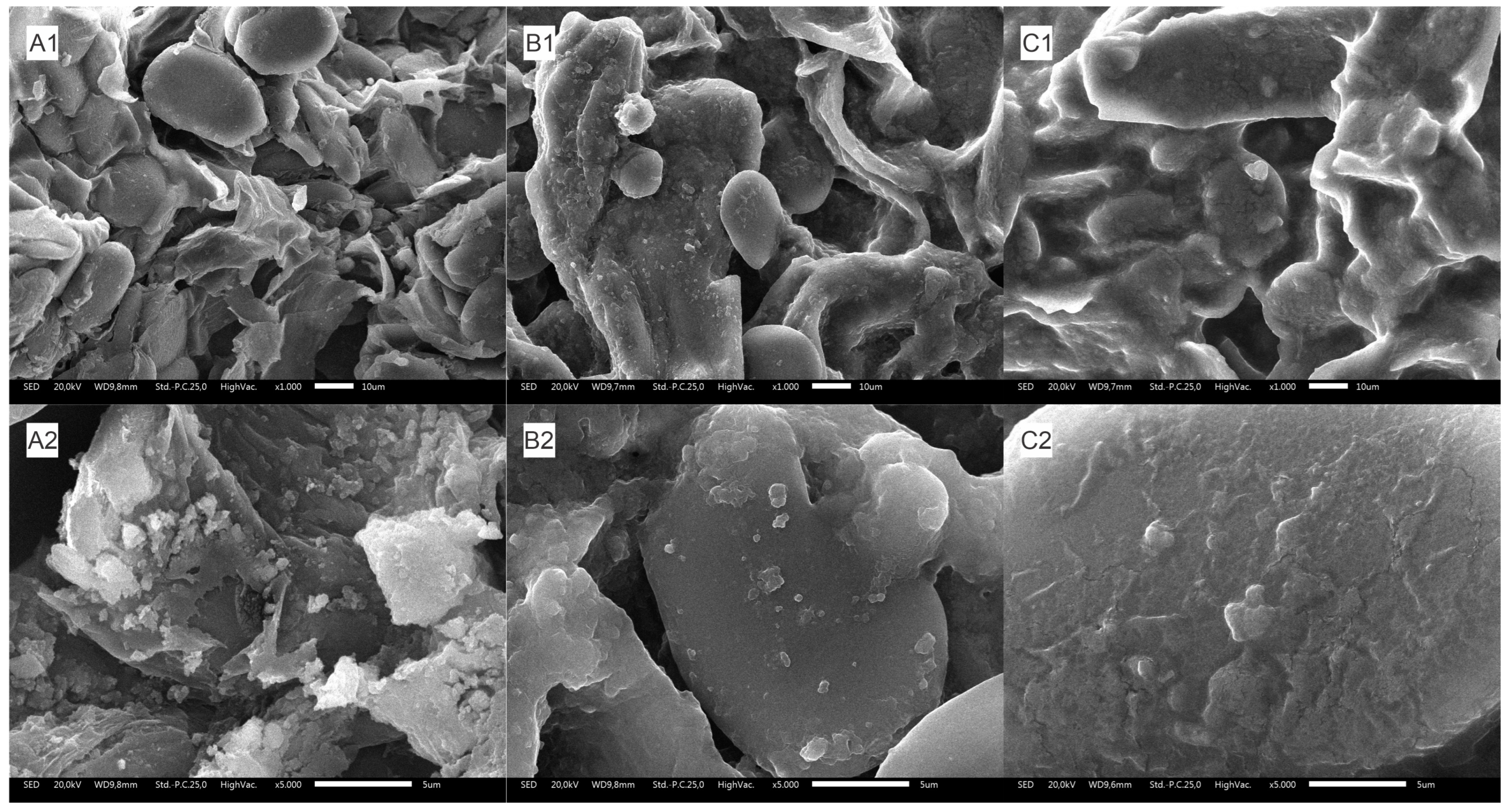
2.5.2. Surface Morphology Analysis
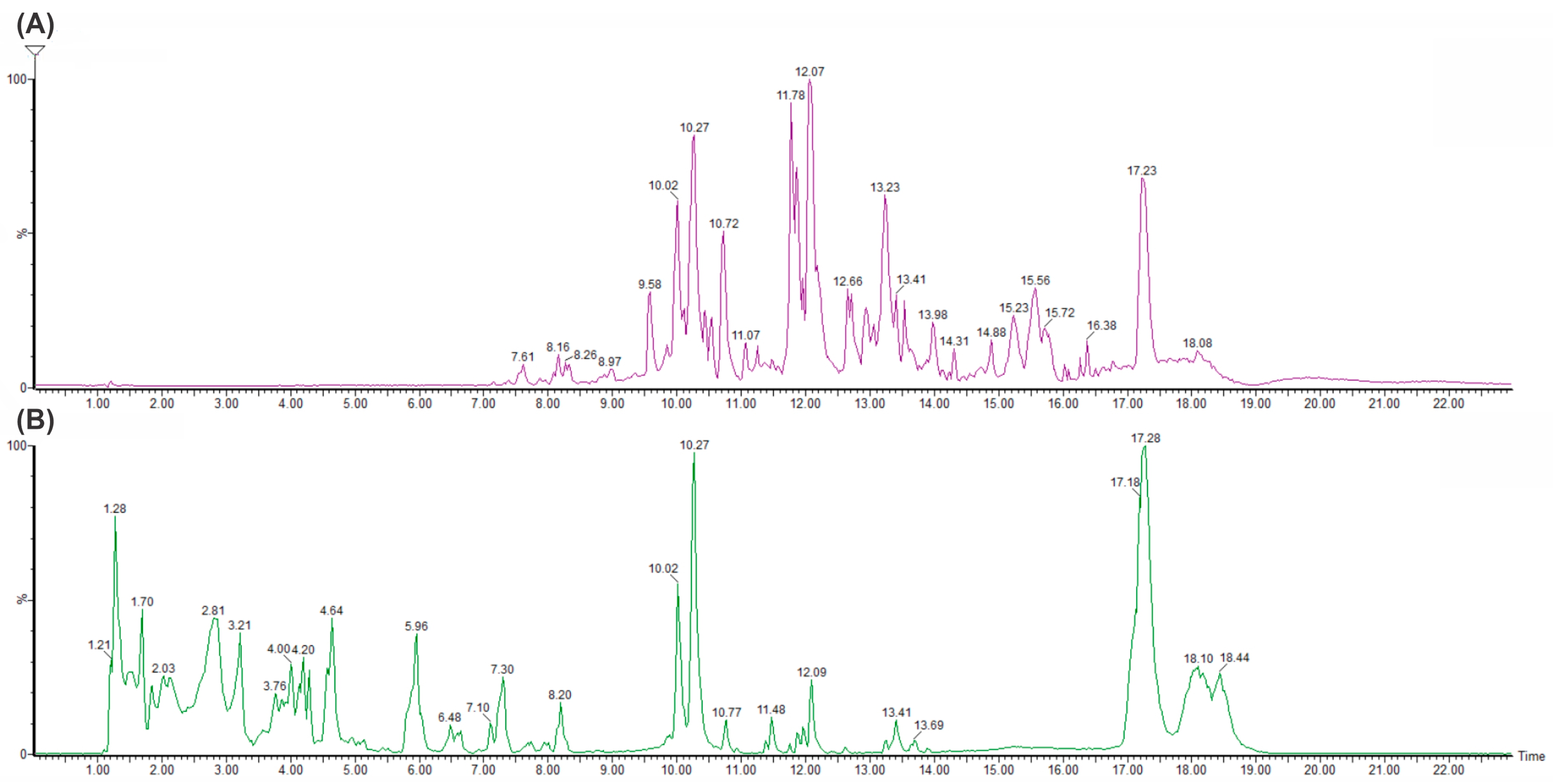
2.5.3. Metabolite Identification of GluLA and Ethanol Extracts
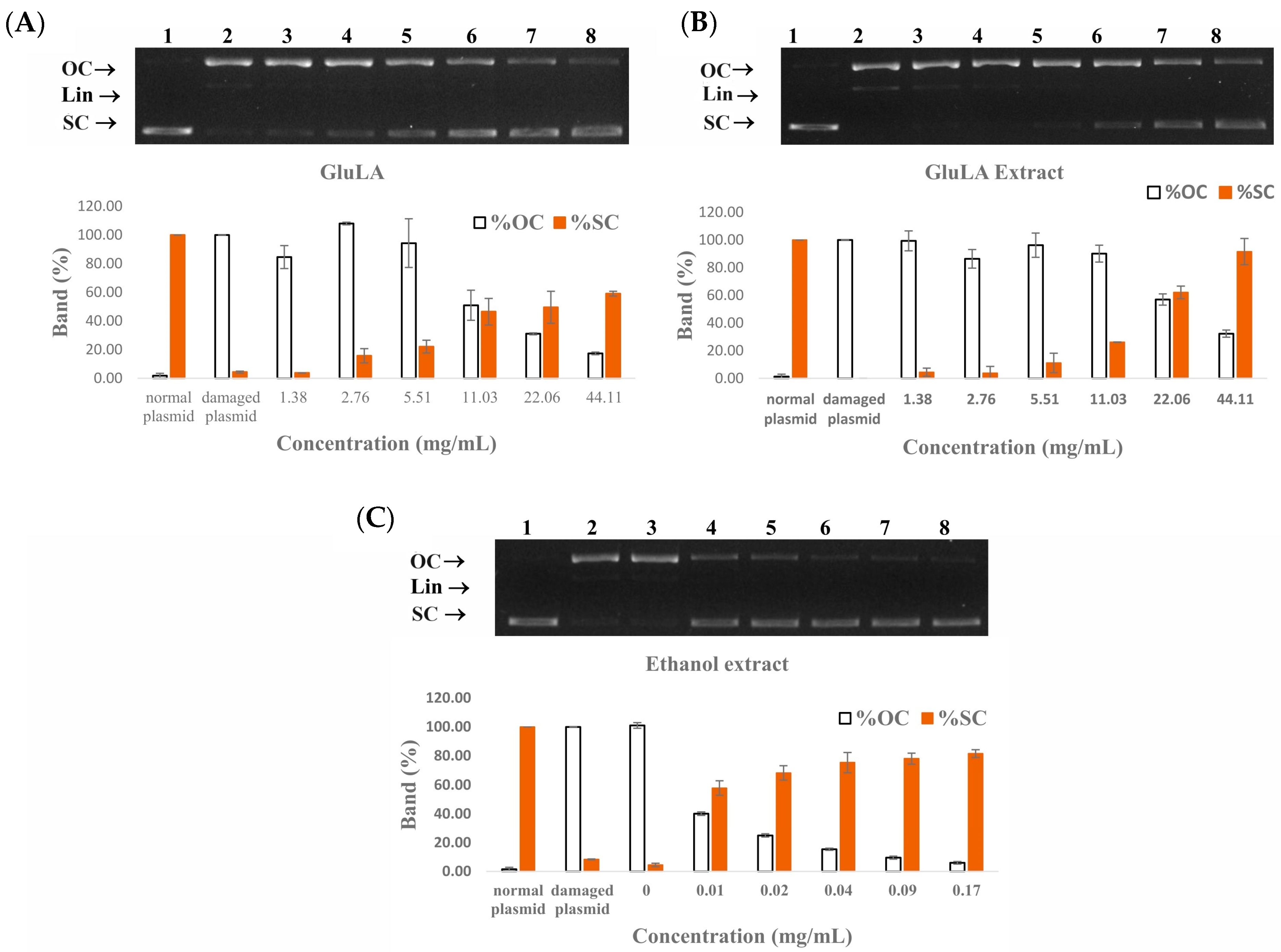
2.5.4. DNA Damage Protection Activity of GluLA and Ethanol Extracts
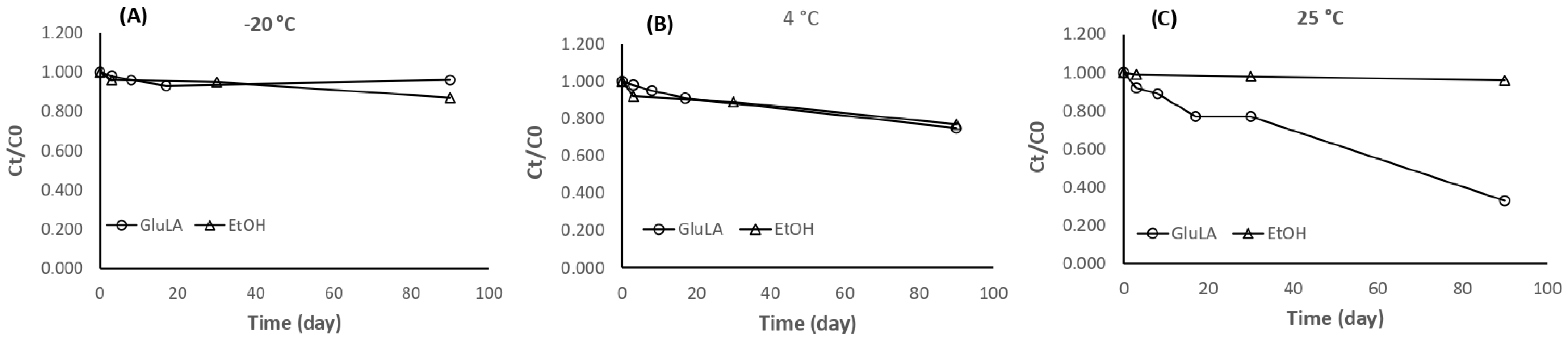
2.5.5. Stability of Xanthorrhizol Extracted by GluLA and Ethanol
2.5.6. FTIR and NMR Characterization of the Optimal NADESs and Interaction of NADESs with Xanthorrhizol
3. Materials and Methods
3.1. Plant Material and Chemical Reagents
3.2. NADES Preparation
3.3. Physical Properties of NADESs
3.4. Xanthorrhizol Extraction
3.5. Extraction Optimization Using Response Surface Methodology
3.6. Determination of Xanthorrhizol Content by TLC Densitometric Analysis
3.7. Phytochemical Analysis by UPLC-QTOF-MS
3.8. Determination of Antioxidant Activity
3.8.1. DPPH Radical-Scavenging Activity Assay
3.8.2. Ferric Reducing/Antioxidant Power Activity Assay
3.9. DNA Protection Assay
3.10. Real-Time Bacterial Growth Determination
3.11. FTIR Spectroscopy Analysis
3.12. NMR Spectroscopy Analysis
3.13. Surface Morphology Characterization
3.14. Storage Stability Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirschhorn, H.H. Botanical remedies of the former Dutch East Indies (Indonesia). Part II: Dicotyledones up to and including leguminosae. J. Ethnopharmacol. 1983, 8, 65–96. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Jung, Y.-J.; Antar Aziz Mohamed, M.; Hoon Lee, T.; Lee, C.-H.; Han, D.; Song, M.-C.; Kim, J.; Baek, N.-I. New bisabolane sesquiterpenes from the rhizomes of Curcuma xanthorrhiza Roxb. and their inhibitory effects on UVB-induced MMP-1 expression in human keratinocytes. Helv. Chim. Acta. 2014, 97, 438–446. [Google Scholar] [CrossRef]
- Halim, M.R.A.; Tan, M.; Ismail, S.; Mahmud, R. Standardization and phytochemical studies of Curcuma xanthorrhiza Roxb. Int. J. Pharm. Pharm. Sci. 2012, 4, 606–610. [Google Scholar]
- Simamora, A.; Timotius, K.H.; Yerer, M.B.; Setiawan, H.; Mun’im, A. Xanthorrhizol, a potential anticancer agent, from Curcuma xanthorrhiza Roxb. Phytomedicine 2022, 105, 154359. [Google Scholar] [CrossRef] [PubMed]
- Akarchariya, N.; Sirilun, S.; Julsrigival, J.; Chansakaowa, S. Chemical profiling and antimicrobial activity of essential oil from Curcuma aeruginosa Roxb., Curcuma glans K. Larsen & J. Mood and Curcuma cf. xanthorrhiza Roxb. collected in Thailand. Asian Pac. J. Trop. Biomed. 2017, 7, 881–885. [Google Scholar] [CrossRef]
- Kim, M.B.; Kim, C.; Song, Y.; Hwang, J.K. Antihyperglycemic and anti-inflammatory effects of standardized Curcuma xanthorrhiza Roxb. Extract and its active compound xanthorrhizol in high-fat diet-induced obese mice. Evid. Based Complement. Altern. Med. 2014, 2014, 205915. [Google Scholar] [CrossRef]
- Lim, C.S.; Jin, D.-Q.; Mok, H.; Oh, S.J.; Lee, J.U.; Hwang, J.K.; Ha, I.; Han, J.-S. Antioxidant and antiinflammatory activities of xanthorrhizol in hippocampal neurons and primary cultured microglia. J. Neurosci. Res. 2005, 82, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Erpina, E.; Rafi, M.; Darusman, L.K.; Vitasari, A.; Putra, B.R.; Rohaeti, E. Simultaneous quantification of curcuminoids and xanthorrhizol in Curcuma xanthorrhiza by high-performance liquid chromatography. J. Liq. Chromatogr. Rel. Technol. 2017, 40, 635–639. [Google Scholar] [CrossRef]
- Altunay, N.; Elik, A.; Gürkan, R. Preparation and application of alcohol based deep eutectic solvents for extraction of curcumin in food samples prior to its spectrophotometric determination. Food Chem. 2020, 310, 125933. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Fu, R.; Zhang, L.; Wang, D.; Wang, S. Enhanced extraction of natural pigments from Curcuma longa L. using natural deep eutectic solvents. Ind. Crops Prod. 2019, 140, 111620. [Google Scholar] [CrossRef]
- Ramdani, E.D.; Marlupi, U.D.; Sinambela, J.; Tjandrawinata, R.R. A new method of xanthorrhizol isolation from the rhizome extract of Curcuma xanthorrhiza. Sch. Acad. J. Biosci. 2016, 4, 732–737. [Google Scholar] [CrossRef]
- Abbott, A.P.; Boothby, D.; Capper, G.; Davies, D.L.; Rasheed, R.K. Deep eutectic solvents formed between choline chloride and carboxylic acids: Versatile alternatives to ionic liquids. J. Am. Chem. Soc. 2004, 126, 9142–9147. [Google Scholar] [CrossRef]
- Oktaviyanti, N.D.; Kartini, K.; Hadiyat, M.A.; Rachmawati, E.; Wijaya, A.C.; Hayun, H.; Mun’im, A. A green extraction design for enhancing flavonoid compounds from Ixora javanica flowers using a deep eutectic solvent. R. Soc. Open Sci. 2020, 7, 201116. [Google Scholar] [CrossRef]
- Zannou, O.; Pashazadeh, H.; Ghellam, M.; Ibrahim, S.A.; Koca, I. Extraction of anthocyanins from borage (Echium amoenum) flowers using choline chloride and a glycerol-based, deep eutectic solvent: Optimization, antioxidant activity, and in vitro bioavailability. Molecules 2022, 27, 134. [Google Scholar] [CrossRef]
- Koigerova, A.; Gosteva, A.; Samarov, A.; Tsvetov, N. Deep Eutectic Solvents Based on Carboxylic Acids and Glycerol or Propylene Glycol as Green Media for Extraction of Bioactive Substances from Chamaenerion angustifolium (L.) Scop. Molecules 2023, 28, 6978. [Google Scholar] [CrossRef]
- Petrochenko, A.A.; Orlova, A.; Frolova, N.; Serebryakov, E.B.; Soboleva, A.; Flisyuk, E.V.; Frolov, A.; Shikov, A.N. Natural Deep Eutectic Solvents for the Extraction of Triterpene Saponins from Aralia elata var. mandshurica (Rupr. & Maxim.) J. Wen. Molecules 2023, 28, 3614. [Google Scholar] [CrossRef] [PubMed]
- Craveiro, R.; Aroso, I.; Flammia, V.; Carvalho, T.; Viciosa, M.T.; Dionísio, M.; Barreiros, S.; Reis, R.L.; Duarte, A.R.C.; Paiva, A. Properties and thermal behavior of natural deep eutectic solvents. J. Mol. Liq. 2016, 215, 534–540. [Google Scholar] [CrossRef]
- Mitar, A.; Panić, M.; Prlić Kardum, J.; Halambek, J.; Sander, A.; Zagajski Kučan, K.; Radojčić Redovniković, I.; Radošević, K. Physicochemical properties, cytotoxicity, and antioxidative activity of natural deep eutectic solvents containing organic acid. Chem. Biochem. Eng. Q. 2019, 33, 1–18. [Google Scholar] [CrossRef]
- Jurić, T.; Uka, D.; Holló, B.B.; Jović, B.; Kordić, B.; Popović, B.M. Comprehensive physicochemical evaluation of choline chloride-based natural deep eutectic solvents. J. Mol. Liq. 2021, 343, 116968. [Google Scholar] [CrossRef]
- Patil, S.S.; Pathak, A.; Rathod, V.K. Optimization and kinetic study of ultrasound assisted deep eutectic solvent based extraction: A greener route for extraction of curcuminoids from Curcuma longa. Ultrason. Sonochem. 2021, 70, 105267. [Google Scholar] [CrossRef]
- Rente, D.; Paiva, A.; Duarte, A.R. The Role of hydrogen bond donor on the extraction of phenolic compounds from natural matrices using deep eutectic systems. Molecules 2021, 26, 2336. [Google Scholar] [CrossRef]
- Nam, M.W.; Zhao, J.; Lee, M.S.; Jeong, J.H.; Lee, J. Enhanced extraction of bioactive natural products using tailor-made deep eutectic solvents: Application to flavonoid extraction from Flos sophorae. Green Chem. 2015, 17, 1718–1727. [Google Scholar] [CrossRef]
- Hayyan, M.; Looi, C.Y.; Hayyan, A.; Wong, W.F.; Hashim, M.A. In vitro and in vivo toxicity profiling of ammonium-based deep eutectic solvents. PLoS ONE 2015, 10, e0117934. [Google Scholar] [CrossRef] [PubMed]
- Hayyan, A.; Mjalli, F.S.; AlNashef, I.M.; Al-Wahaibi, Y.M.; Al-Wahaibi, T.; Hashim, M.A. Glucose-based deep eutectic solvents: Physical properties. J. Mol. Liq. 2013, 178, 137–141. [Google Scholar] [CrossRef]
- Lomba, L.; Ribate, M.; Sangüesa, E.; Concha, J.; Garralaga, M.; Errazquin, D.; García, C.B.; Giner, B. Deep eutectic solvents: Are they safe? Appl. Sci. 2021, 11, 10061. [Google Scholar] [CrossRef]
- Torregrosa-Crespo, J.; Marset, X.; Guillena, G.; Ramón, D.J.; María Martínez-Espinosa, R. New guidelines for testing “Deep eutectic solvents” toxicity and their effects on the environment and living beings. Sci. Total Environ. 2020, 704, 135382. [Google Scholar] [CrossRef] [PubMed]
- Clifton, L.A.; Skoda, M.W.A.; Le Brun, A.P.; Ciesielski, F.; Kuzmenko, I.; Holt, S.A.; Lakey, J.H. Effect of divalent cation removal on the structure of Gram-negative bacterial outer membrane models. Langmuir 2015, 31, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Lund, P.A.; De Biase, D.; Liran, O.; Scheler, O.; Mira, N.P.; Cetecioglu, Z.; Fernández, E.N.; Bover-Cid, S.; Hall, R.; Sauer, M.; et al. Understanding how microorganisms respond to acid pH is central to their control and successful exploitation. Front. Microbiol. 2020, 11, 556140. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, J.; Zhang, H.; Dzah, C.S.; Zandile, M.; Duan, Y.; Ma, H.; Luo, X. Advances in ultrasound assisted extraction of bioactive compounds from cash crops—A review. Ultrason. Sonochem. 2018, 48, 538–549. [Google Scholar] [CrossRef]
- Klau, M.E.; Rohaeti, E.; Rafi, M.; Artika, I.M.; Ambarsari, L.; Nurcholis, W. Metabolite profiling of Curcuma xanthorriza varieties grown in different regions using UHPLC-Q-Orbitrap-HRMS and chemometrics analysis. Biointerface Res. Appl. Chem. 2023, 13, 1–13. [Google Scholar] [CrossRef]
- Zhang, C.-M.; Fan, P.-H.; Li, M.; Lou, H.-X. Two new sesquiterpenoids from the rhizomes of Curcuma xanthorrhiza. Helv. Chim. Acta. 2014, 97, 1295–1300. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, J.; Ji, M.; Fan, P. Acetylcholinesterase inhibitors and compounds promoting SIRT1 expression from Curcuma xanthorrhiza. Phytochem. Lett. 2015, 12, 215–219. [Google Scholar] [CrossRef]
- Huang, H.; Zhu, Y.; Fu, X.; Zou, Y.; Li, Q.; Luo, Z. Integrated natural deep eutectic solvent and pulse-ultrasonication for efficient extraction of crocins from gardenia fruits (Gardenia jasminoides Ellis) and its bioactivities. Food Chem. 2022, 380, 132216. [Google Scholar] [CrossRef]
- Xu, J.-G.; Hu, Q.-P.; Liu, Y. Antioxidant and DNA-protective activities of chlorogenic acid isomers. J. Agric. Food Chem. 2012, 60, 11625–11630. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; van Spronsen, J.; Witkamp, G.J.; Verpoorte, R.; Choi, Y.H. Natural deep eutectic solvents as new potential media for green technology. Anal. Chim. Acta 2013, 766, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Verpoorte, R.; Choi, Y.H. Natural deep eutectic solvents providing enhanced stability of natural colorants from safflower (Carthamus tinctorius). Food Chem. 2014, 159, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Abou-Taleb, N.H.; El-Sherbiny, D.T.; El-Enany, N.M.; El-Subbagh, H.I. A new grey relational analysis application in analytical chemistry: Natural deep eutectic solvent as a green extractant for HPLC determination of lamotrigine in plasma. Microchem. J. 2022, 172, 106918. [Google Scholar] [CrossRef]
- Santana, A.P.R.; Mora-Vargas, J.A.; Guimarães, T.G.S.; Amaral, C.D.B.; Oliveira, A.; Gonzalez, M.H. Sustainable synthesis of natural deep eutectic solvents (NADES) by different methods. J. Mol. Liq. 2019, 293, 111452. [Google Scholar] [CrossRef]
- Duan, L.; Dou, L.-L.; Guo, L.; Li, P.; Liu, E.H. Comprehensive Evaluation of Deep Eutectic Solvents in Extraction of Bioactive Natural Products. ACS Sustain. Chem. Eng. 2016, 4, 2405–2411. [Google Scholar] [CrossRef]
- Saputri, F.A.; Mun’im, A.; Putri, C.R.; Aryani, D. Validasi metode analisis kurkuminoid dan xantorizol pada rimpang temulawak (Curcuma xanthorrhiza) dengan KLT-densitometri. Media Pharm. Indones. 2022, 4, 147–156. [Google Scholar] [CrossRef]
- Hilma, R.; Wulandari, E.T.S.; Arman, Z. Fruit stalk extract from chili peppers (Capsicum annum L.) as a natural antioxidant to inhibit oxidation in crude palm oil. J. Kim. Sains Dan Apl. 2020, 23, 124–128. [Google Scholar] [CrossRef]
- Jimenez-Alvarez, D.; Giuffrida, F.; Vanrobaeys, F.; Golay, P.A.; Cotting, C.; Lardeau, A.; Keely, B.J. High-throughput methods to assess lipophilic and hydrophilic antioxidant capacity of food extracts in vitro. J. Agric. Food Chem. 2008, 56, 3470–3477. [Google Scholar] [CrossRef] [PubMed]
- Rahayu, I.; Timotius, K.H. Phytochemical analysis, antimutagenic and antiviral activity of Moringa oleifera L. leaf infusion: In vitro and in silico studies. Molecules 2022, 27, 4017. [Google Scholar] [CrossRef] [PubMed]
- Simamora, A.; Timotius, K.H.; Setiawan, H.; Putra, M.Y.; Mun’im, A. Natural deep eutectic solvent extraction of xanthorrhizol and curcuminoids from Curcuma xanthorrhiza Roxb and simultaneous determination by high-performance liquid chromatography. J. Pharm. Pharmacogn. Res. 2023, 11, 1056–1071. [Google Scholar] [CrossRef]
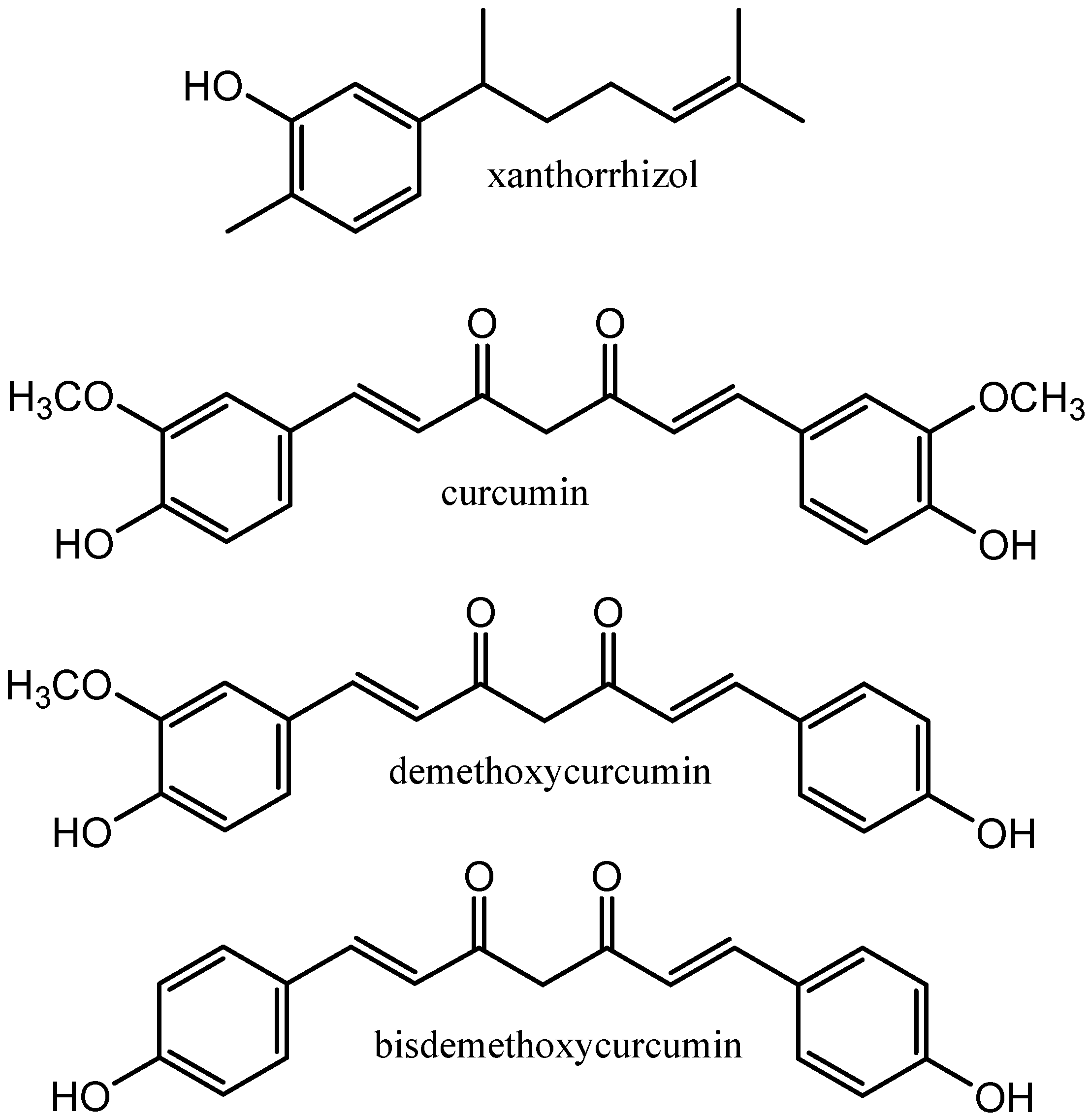
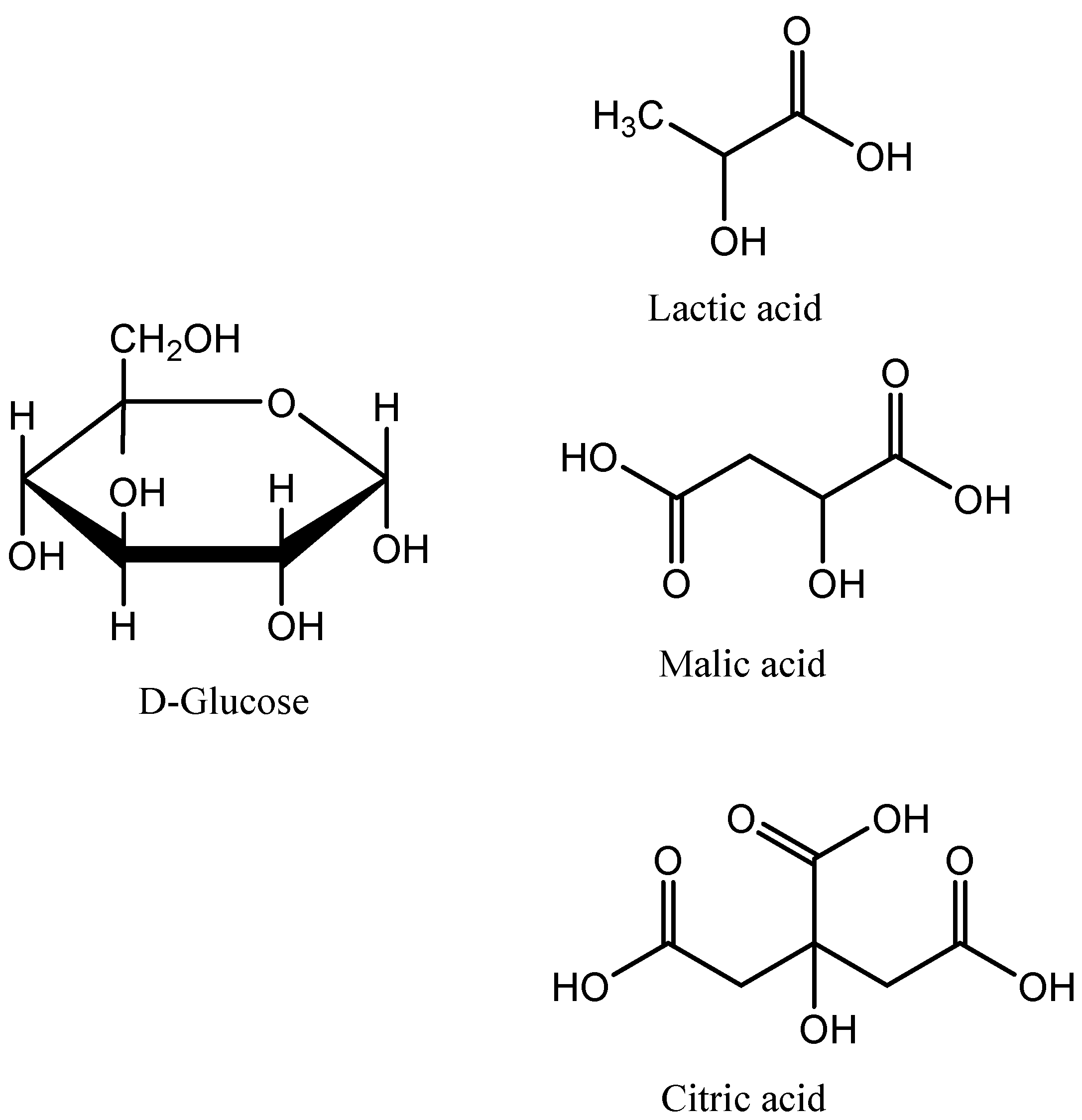




| Sample Name | Polarity (kJ mol−1) | Viscosity (mPa s) | Density (kg m−3) | FRAP (µG TE g−1 NADESs or Solvent) * | DPPH (µG AAE g−1 NADESs or Solvent) ** |
|---|---|---|---|---|---|
| GluLA | 198.70 | 113.9 | 1.31 | 29.12 ± 0.09 | 50.42 ± 0.50 |
| GluMA | 199.03 | NM § | 1.40 | 9.80 ± 0.13 | 59.53 ± 0.42 |
| GluCA | 207.07 | NM § | 1.46 | 31.30 ± 0.97 | 60.69 ± 2.30 |
| Ethanol | 218.07 | ND # | 0.79 | 14.22 ± 0.32 | 6.53 ± 1.93 |
| Run | Independent Variables | Responses | |||
|---|---|---|---|---|---|
| X1 | X2 | X3 | Xanthorrhizol Content (mg/g) | Curcuminoid Content (mg/g) | |
| 1 | 30 | 15 | 20 | 17.62 | 6.64 |
| 2 | 10 | 15 | 20 | 4.99 | 6.02 |
| 3 | 20 | 10 | 20 | 11.85 | 4.69 |
| 4 | 20 | 15 | 10 | 7.82 | 6.50 |
| 5 | 30 | 5 | 20 | 1.59 | 2.28 |
| 6 | 20 | 15 | 30 | 3.53 | 5.96 |
| 7 | 20 | 5 | 10 | 0 | 2.39 |
| 8 | 30 | 10 | 10 | 6.03 | 4.37 |
| 9 | 20 | 10 | 20 | 11.58 | 4.67 |
| 10 | 10 | 10 | 30 | 2.50 | 4.81 |
| 11 | 10 | 5 | 20 | 9.50 | 3.46 |
| 12 | 20 | 10 | 20 | 11.98 | 4.61 |
| 13 | 30 | 10 | 30 | 5.07 | 4.46 |
| 14 | 20 | 10 | 20 | 11.99 | 4.58 |
| 15 | 10 | 10 | 10 | 5.50 | 4.42 |
| 16 | 20 | 5 | 30 | 0 | 3.37 |
| 17 | 20 | 10 | 20 | 11.90 | 4.64 |
| Identification | Category | RT (min) | Formula | Measured Mass (Da) | Calculated Mass (Da) | Error (ppm) | MS Fragmentation (m/z) |
|---|---|---|---|---|---|---|---|
| Ethanol extract | |||||||
| Demethoxycurcumin | Diarylheptanoids | 10.020 | C20H18O5 | 338.1225 | 338.1232 | −2.1 | 339, 177, 147 |
| Curcumin | Diarylheptanoids | 10.267 | C21H20O6 | 368.1338 | 368.1362 | 6.5 | 369, 285, 177, 149 |
| Zedoarol | Terpenoids | 10.723 | C15H18O3 | 268.1151 | 268.1151 | 0 | 269, 247,229 |
| Dihydropyrocurzerenon | Terpenoids | 11.074 | C15H18O | 214.1436 | 214.1436 | 0 | 215, 159 |
| Isovelleral | Terpenoids | 11.777 | C15H18O | 232.1528 | 232.1542 | −6.0 | 233, 215, 197 |
| Curzerenone | Terpenoids | 12.087 | C15H18O2 | 230.1382 | 230.1385 | −1.3 | 231, 213, 149 |
| Curcumene | Terpenoids | 12.706 | C15H22 | 202.1799 | 202.1800 | −0.5 | 203, 147, 119, 91 |
| Xanthorrhizol | Terpenoids | 13.234 | C15H22O | 218.1768 | 218.1749 | 8.7 | 219, 201 |
| α-Farnesene/elemene | Terpenoids | 13.979 | C15H24 | 204.1948 | 204.1956 | −3.9 | 205, 149, 118 |
| GluLA extract | |||||||
| 3-Hydroxy-p-cymene | Terpenoids | 1.106 | C10H14O | 150.0352 | 150.0387 | −23.2 | 151,125, 110 |
| ar-Turmerone | Terpenoids | 4.923 | C10H16O5 | 217.1080 | 217.1076 | 1.8 | 217, 145, 128 |
| Demethoxycurcumin | Diarylheptanoids | 9.936 | C20H18O5 | 338.1241 | 338.1232 | 2.7 | 339, 177, 147 |
| Curcumin | Diarylheptanoids | 10.246 | C21H20O6 | 368.1333 | 368.1339 | −1.4 | 369, 285, 177, 147 |
| Gweicurculactone | Lactone | 12.024 | C20H26O2 | 298.1985 | 2 | −8.7 | 299, 253, 155 |
| Salvinolone | Phenanthrene | 12.200 | C20H26O3 | 314.1931 | 314.1960 | −9.2 | 315, 187 |
| Xanthorrhizol | Terpenoids | 13.234 | C15H22O | 218.1742 | 218.1749 | −3.2 | 219, 201, 159, 145 |
| 4’-Hydroxy-5,7-dimethoxyflavanone | Flavanone | 13.409 | C17H16O5 | 300.1788 | 300.1804 | −5.3 | 301, 231, 213 |
| Salvigenin | Flavone | 13.937 | C18H16O6 | 328.2126 | 328.2117 | 2.7 | 329, 173, 131 |
| Stigmasterol | Steroids | 18.415 | C29H48O | 412.3792 | 412.3783 | 2.2 | 429, 413, 139 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simamora, A.; Timotius, K.H.; Setiawan, H.; Saputri, F.A.; Putri, C.R.; Aryani, D.; Ningrum, R.A.; Mun’im, A. Ultrasonic-Assisted Extraction of Xanthorrhizol from Curcuma xanthorrhiza Roxb. Rhizomes by Natural Deep Eutectic Solvents: Optimization, Antioxidant Activity, and Toxicity Profiles. Molecules 2024, 29, 2093. https://doi.org/10.3390/molecules29092093
Simamora A, Timotius KH, Setiawan H, Saputri FA, Putri CR, Aryani D, Ningrum RA, Mun’im A. Ultrasonic-Assisted Extraction of Xanthorrhizol from Curcuma xanthorrhiza Roxb. Rhizomes by Natural Deep Eutectic Solvents: Optimization, Antioxidant Activity, and Toxicity Profiles. Molecules. 2024; 29(9):2093. https://doi.org/10.3390/molecules29092093
Chicago/Turabian StyleSimamora, Adelina, Kris Herawan Timotius, Heri Setiawan, Febrina Amelia Saputri, Chinthia Rahadi Putri, Dewi Aryani, Ratih Asmana Ningrum, and Abdul Mun’im. 2024. "Ultrasonic-Assisted Extraction of Xanthorrhizol from Curcuma xanthorrhiza Roxb. Rhizomes by Natural Deep Eutectic Solvents: Optimization, Antioxidant Activity, and Toxicity Profiles" Molecules 29, no. 9: 2093. https://doi.org/10.3390/molecules29092093
APA StyleSimamora, A., Timotius, K. H., Setiawan, H., Saputri, F. A., Putri, C. R., Aryani, D., Ningrum, R. A., & Mun’im, A. (2024). Ultrasonic-Assisted Extraction of Xanthorrhizol from Curcuma xanthorrhiza Roxb. Rhizomes by Natural Deep Eutectic Solvents: Optimization, Antioxidant Activity, and Toxicity Profiles. Molecules, 29(9), 2093. https://doi.org/10.3390/molecules29092093






