Colored Proteins Act as Biocolorants in Escherichia coli
Abstract
1. Introduction
2. Results
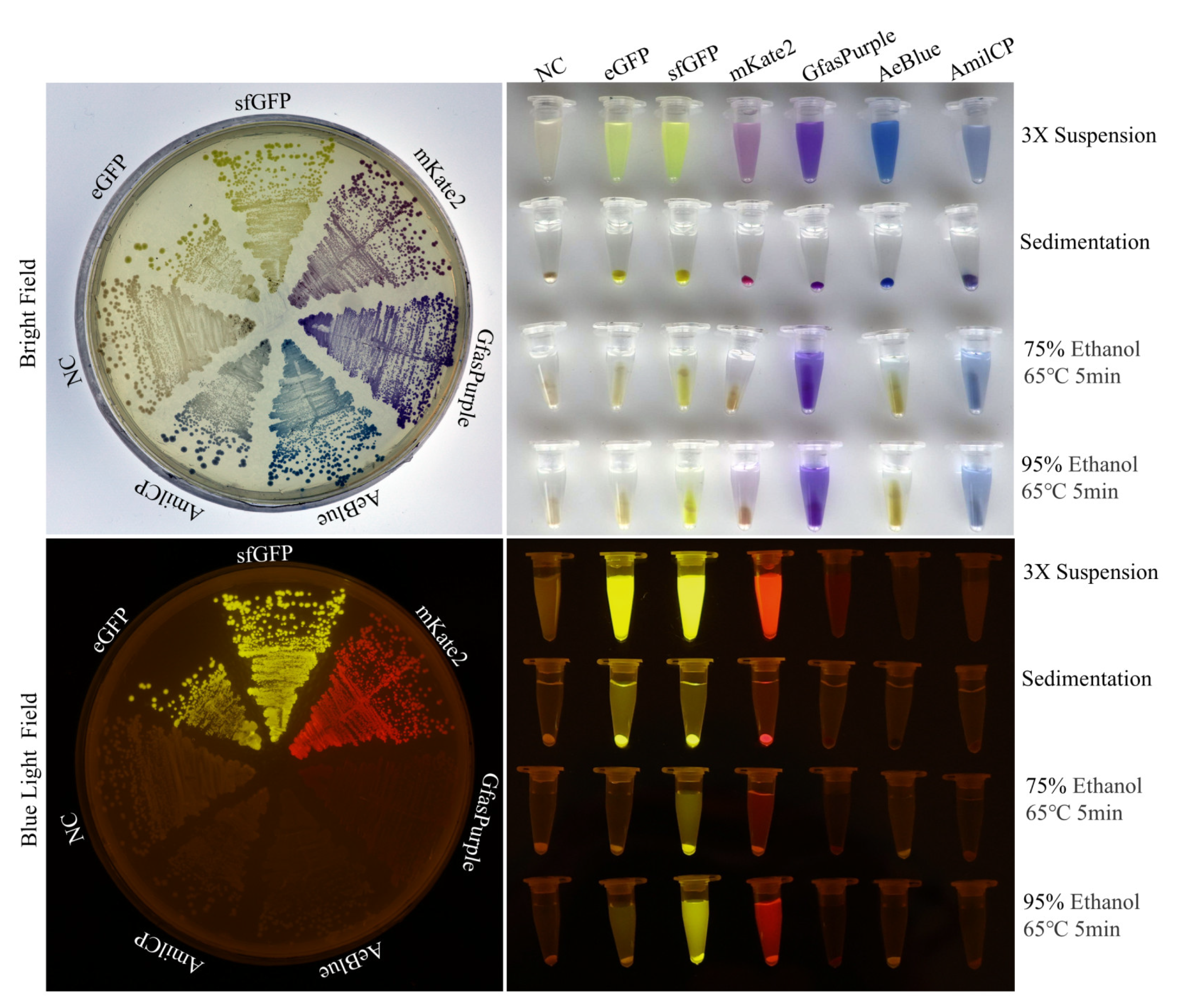
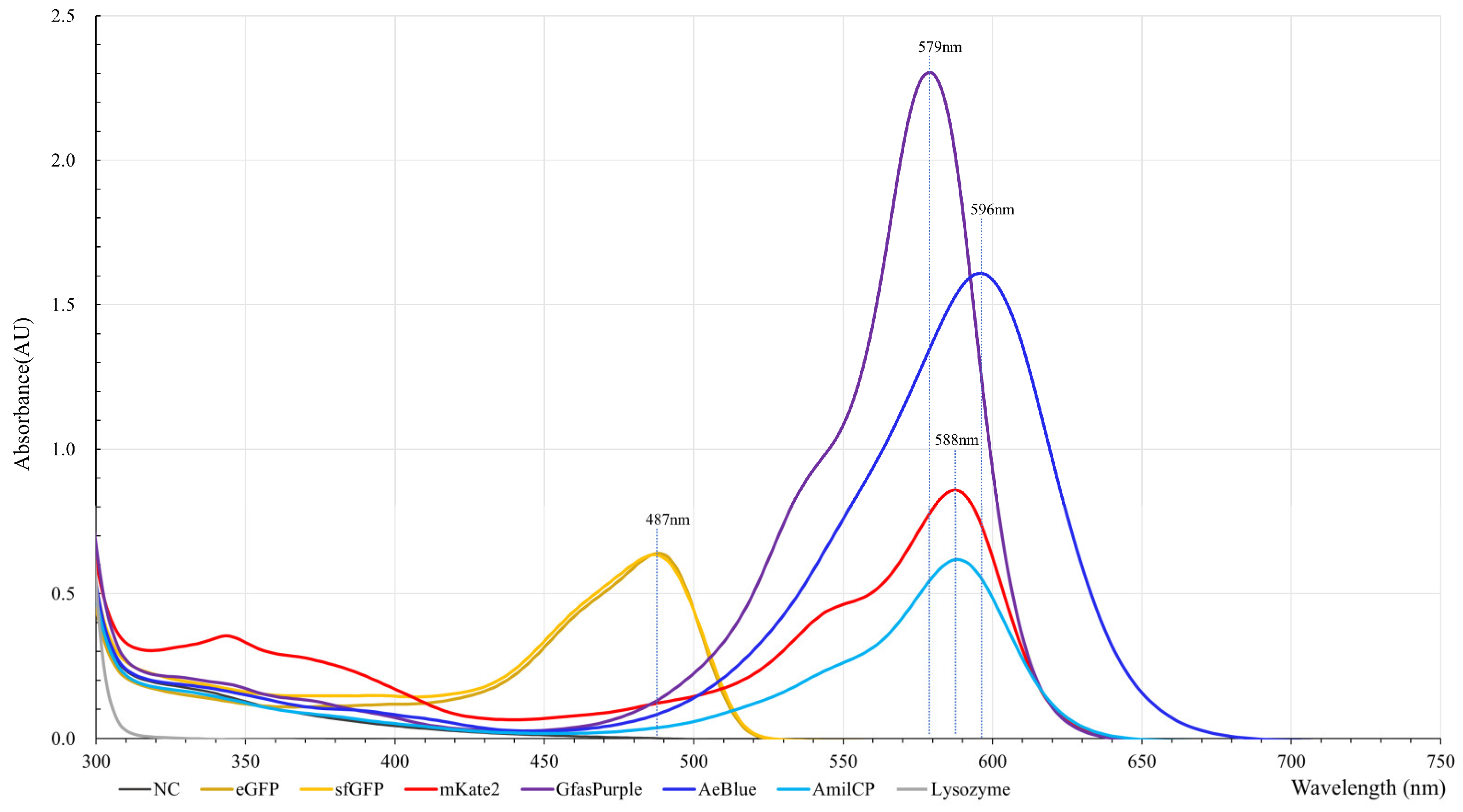
2.1. Production, Extraction, and Spectral Analysis of Colored Proteins
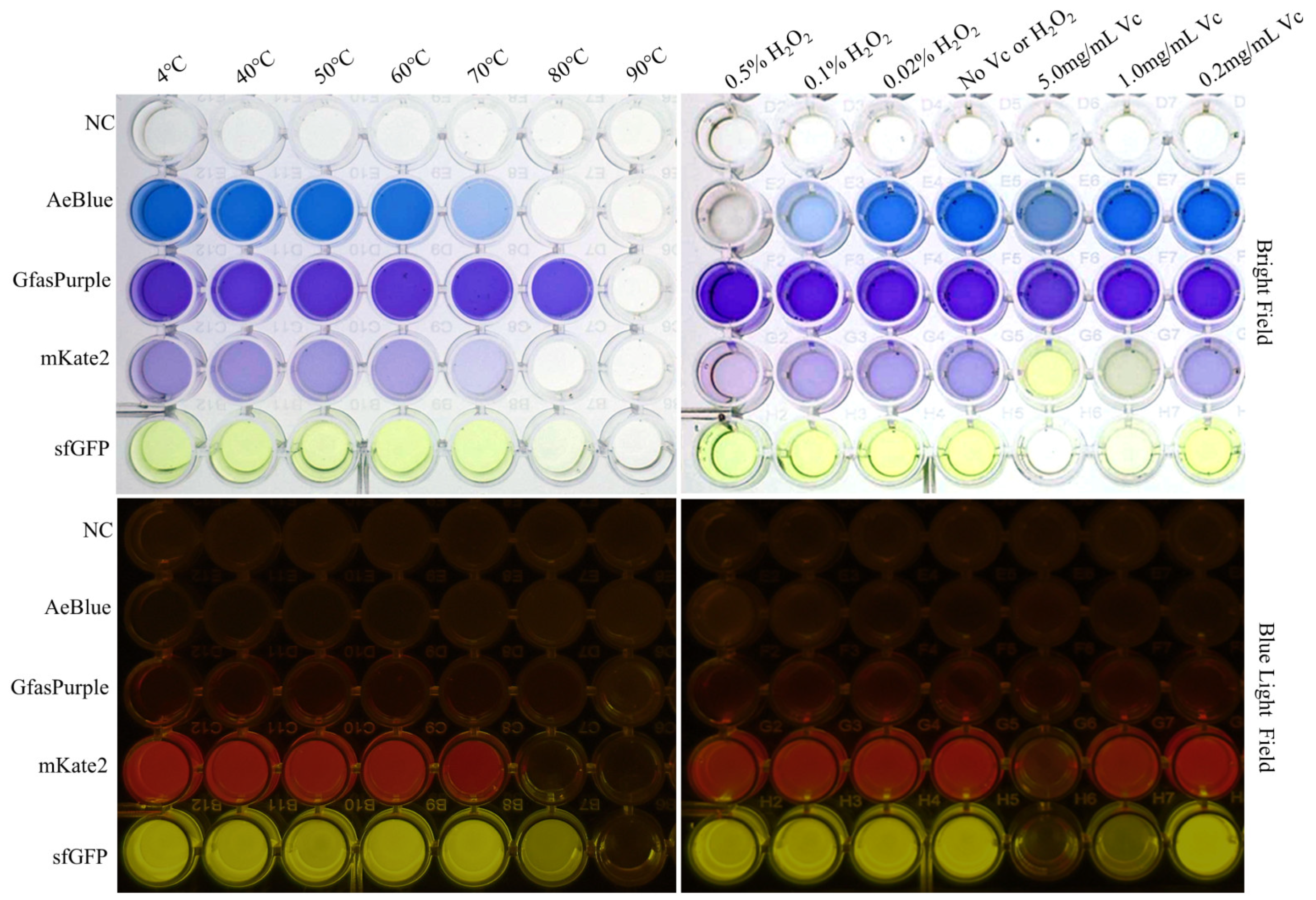
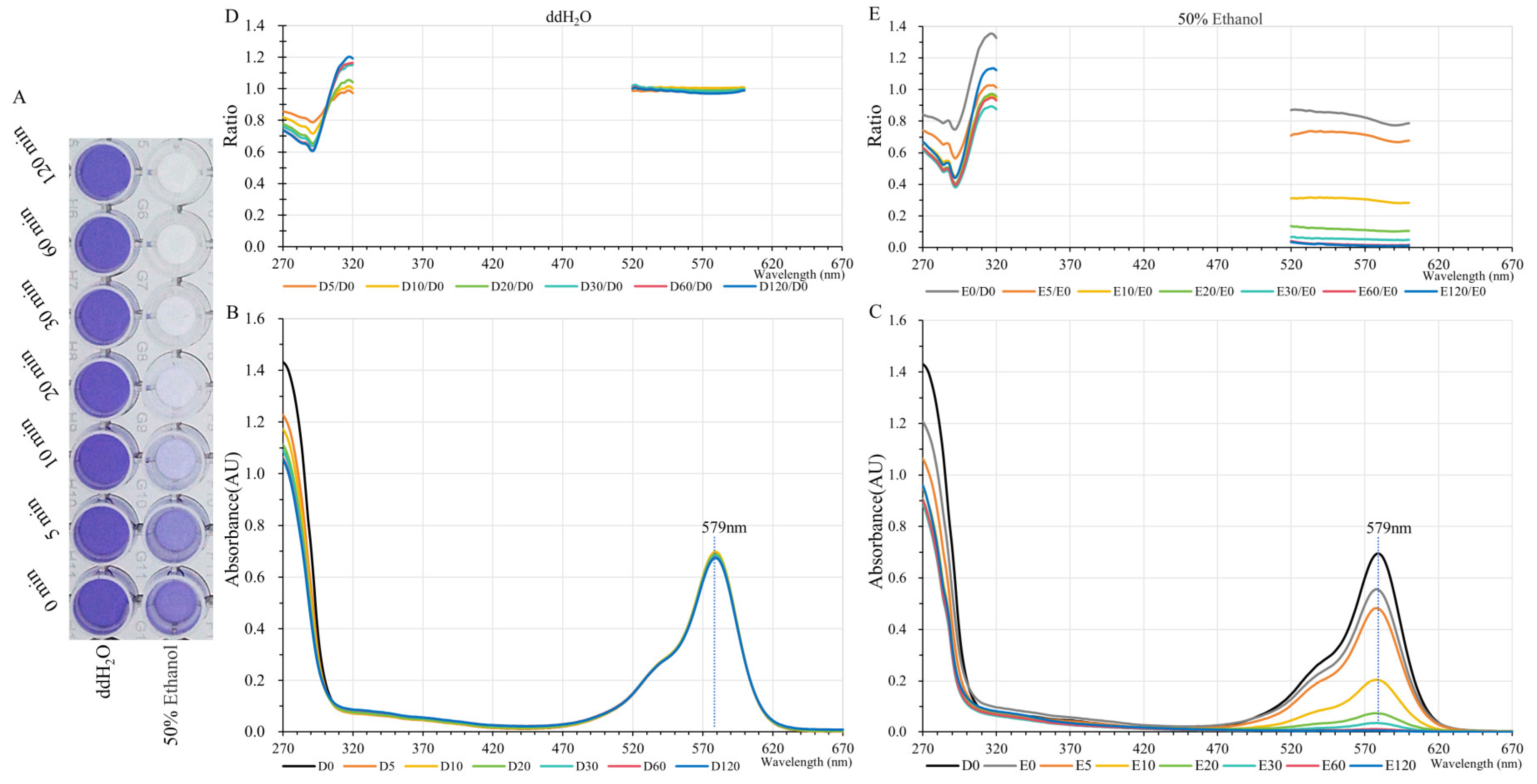
2.2. Heat and Ethanol Tolerance Capabilities of GfasPurple
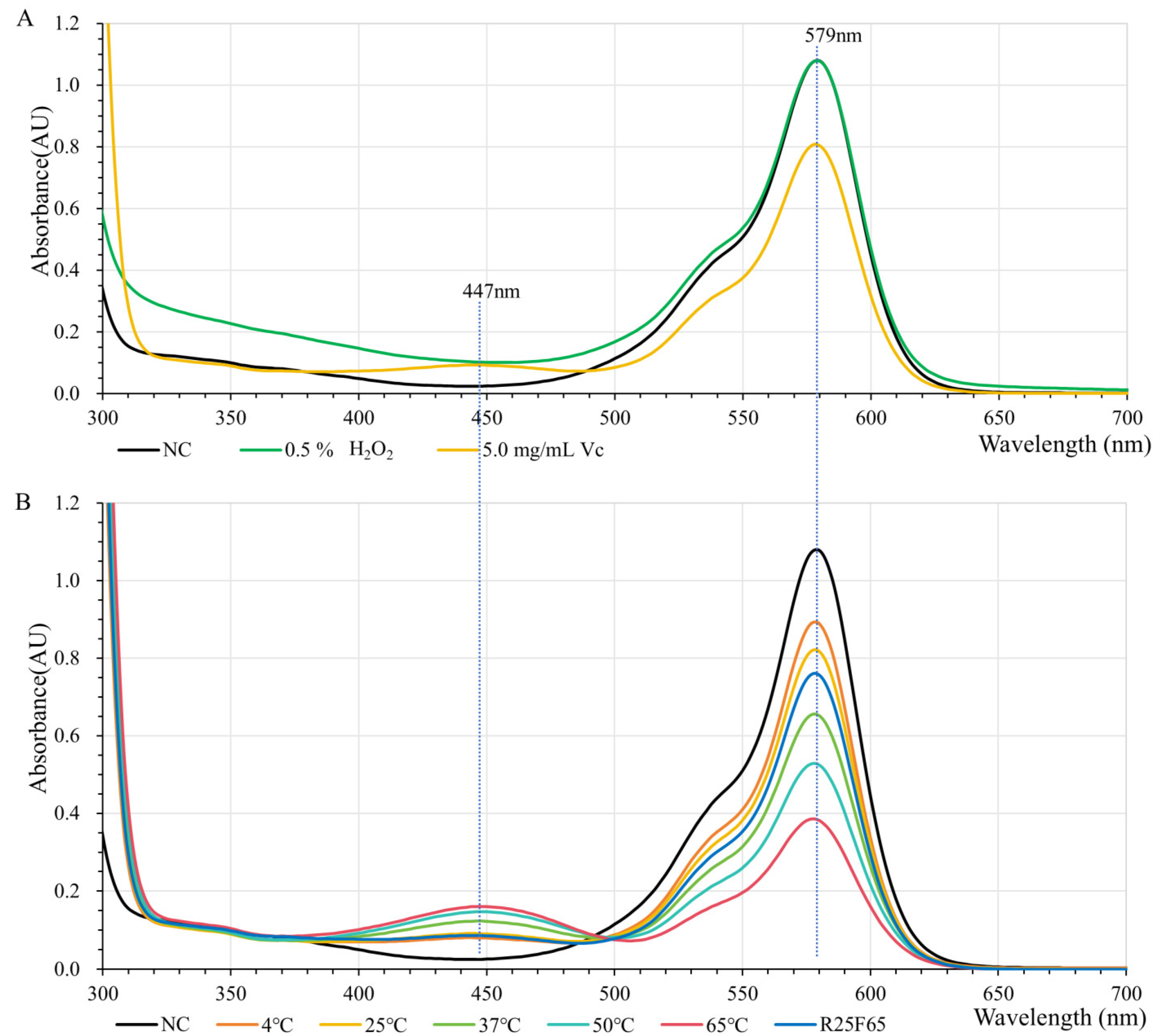
2.3. H2O2 and Vitamin C Tolerance Capabilities of GfasPurple
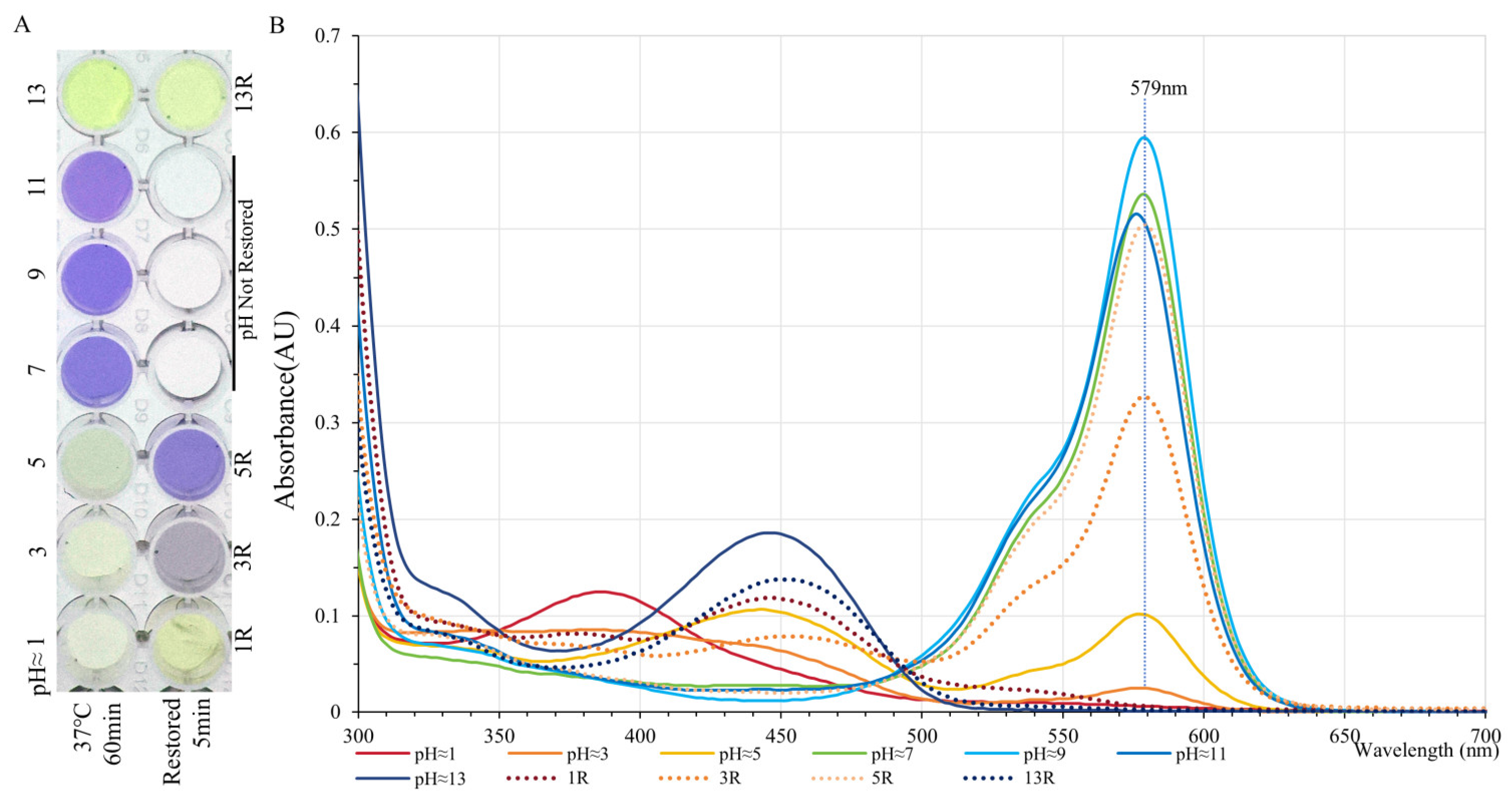
2.4. The Effect of Acidity and Alkalinity on the Color of GfasPurple
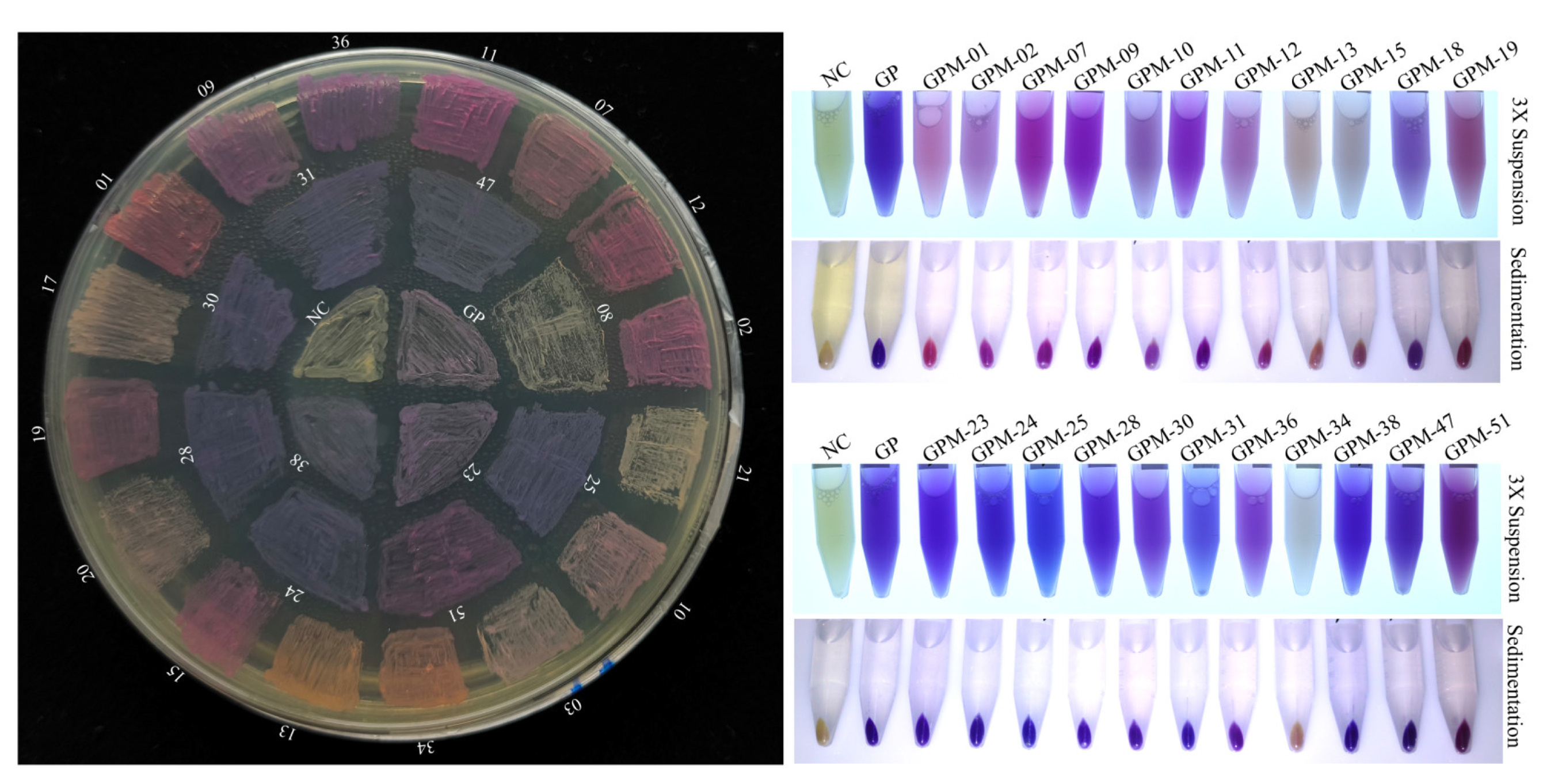
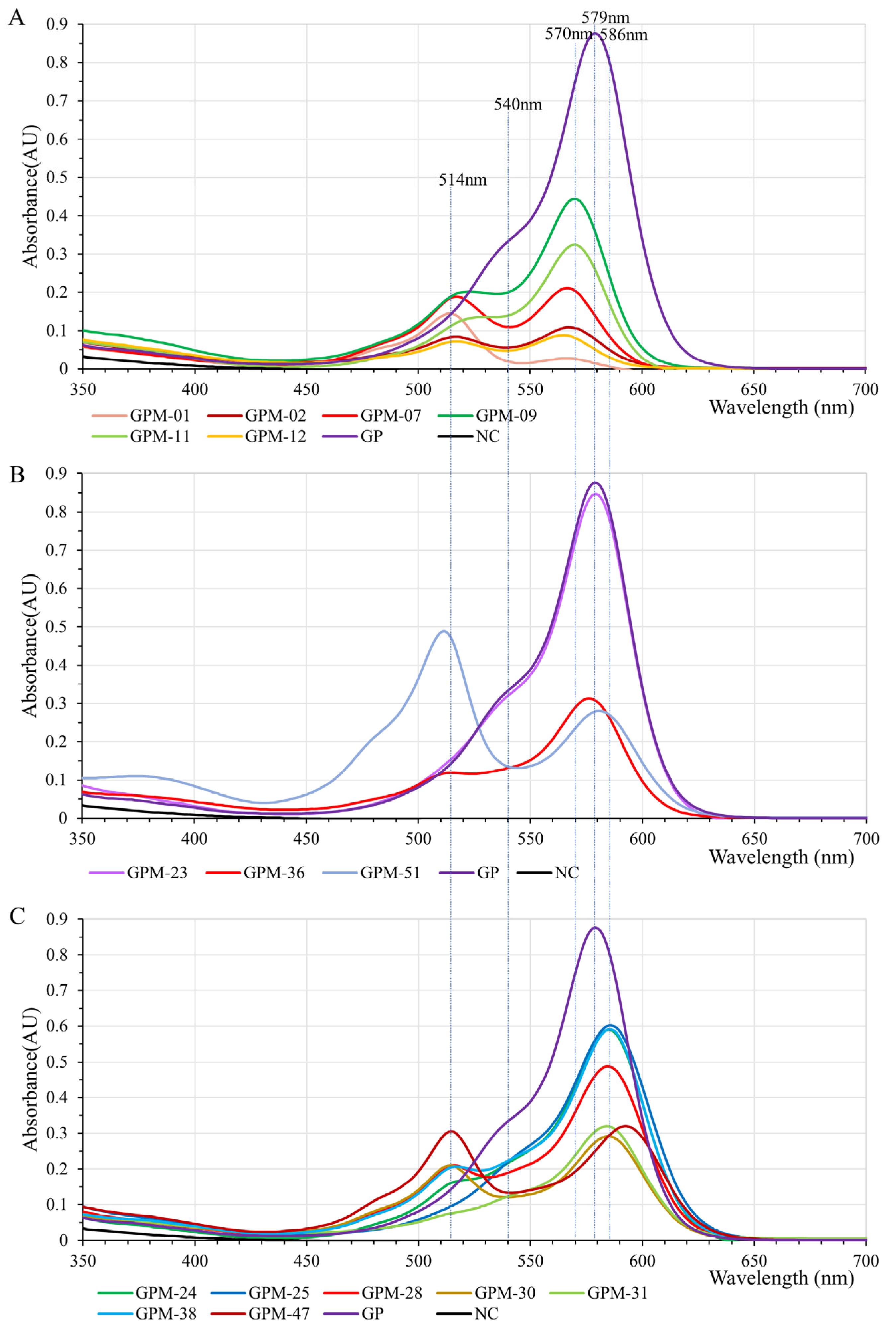
2.5. GfasPurple Variants Display Different Absorption Characteristics in Visible Light Spectrum
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Protein Expression and Extraction
4.3. Absorption Spectroscopy Analysis
4.4. Physicochemical Tests
4.4.1. Heat and Ethanol Tolerance Tests
4.4.2. H2O2 and Vitamin C Tolerance Tests
4.4.3. pH Tests
4.5. GfasPurple Mutagenesis and Screening
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, K.; Beyer, H.M.; Zurbriggen, M.D.; Gartner, W. The Red Edge: Bilin-Binding Photoreceptors as Optogenetic Tools and Fluorescence Reporters. Chem. Rev. 2021, 121, 14906–14956. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Brugliera, F. Flower colour and cytochromes P450. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120432. [Google Scholar] [CrossRef] [PubMed]
- Broser, M. Far-Red Absorbing Rhodopsins, Insights from Heterodimeric Rhodopsin-Cyclases. Front. Mol. Biosci. 2021, 8, 806922. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Wu, Y.; Wang, G.; Jia, T.; Zhang, Y. Purification and bioactivities of phycocyanin. Crit. Rev. Food Sci. Nutr. 2017, 57, 3840–3849. [Google Scholar] [CrossRef] [PubMed]
- Steiner, T.; Hess, P.; Bae, J.H.; Wiltschi, B.; Moroder, L.; Budisa, N. Synthetic biology of proteins: Tuning GFPs folding and stability with fluoroproline. PLoS ONE 2008, 3, e1680. [Google Scholar] [CrossRef]
- Lambert, T.J. FPbase: A community-editable fluorescent protein database. Nat. Methods 2019, 16, 277–278. [Google Scholar] [CrossRef]
- Balleza, E.; Kim, J.M.; Cluzel, P. Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 2018, 15, 47–51. [Google Scholar] [CrossRef]
- Liljeruhm, J.; Funk, S.K.; Tietscher, S.; Edlund, A.D.; Jamal, S.; Wistrand-Yuen, P.; Dyrhage, K.; Gynna, A.; Ivermark, K.; Lovgren, J.; et al. Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. J. Biol. Eng. 2018, 12, 8. [Google Scholar] [CrossRef]
- Shkrob, M.; Mishin, A.S.; Chudakov, D.M.; Labas Iu, A.; Lukyanov, K.A. Chromoproteins of the green fluorescent protein family: Properties and applications. Russ. J. Bioorganic Chem. 2008, 34, 581–590. [Google Scholar] [CrossRef]
- Bao, L.; Menon, P.N.K.; Liljeruhm, J.; Forster, A.C. Overcoming chromoprotein limitations by engineering a red fluorescent protein. Anal. Biochem. 2020, 611, 113936. [Google Scholar] [CrossRef]
- Verkhusha, V.V.; Lukyanov, K.A. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol. 2004, 22, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.Y.; Ma, J.Y.; Li, F.; Zhao, J.R.; Xu, F.C.; Yang, W.W.; Yuan, M.; Gao, W.; Long, L. Optimizing the Protein Fluorescence Reporting System for Somatic Embryogenesis Regeneration Screening and Visual Labeling of Functional Genes in Cotton. Front. Plant Sci. 2021, 12, 825212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Gurtu, V.; Kain, S.R. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem. Biophys. Res. Commun. 1996, 227, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Pedelacq, J.D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef]
- Shkrob, M.A.; Yanushevich, Y.G.; Chudakov, D.M.; Gurskaya, N.G.; Labas, Y.A.; Poponov, S.Y.; Mudrik, N.N.; Lukyanov, S.; Lukyanov, K.A. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochem. J. 2005, 392, 649–654. [Google Scholar] [CrossRef]
- Tamayo-Nunez, J.; de la Mora, J.; Padilla-Vaca, F.; Vargas-Maya, N.I.; Rangel-Serrano, A.; Anaya-Velazquez, F.; Paramo-Perez, I.; Reyes-Martinez, J.E.; Espana-Sanchez, B.L.; Franco, B. aeBlue Chromoprotein Color is Temperature Dependent. Protein Pept. Lett. 2020, 27, 74–84. [Google Scholar] [CrossRef]
- Alieva, N.O.; Konzen, K.A.; Field, S.F.; Meleshkevitch, E.A.; Hunt, M.E.; Beltran-Ramirez, V.; Miller, D.J.; Wiedenmann, J.; Salih, A.; Matz, M.V. Diversity and evolution of coral fluorescent proteins. PLoS ONE 2008, 3, e2680. [Google Scholar] [CrossRef]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef]
- Zhang, C.J.; Mou, H.; Yuan, J.; Wang, Y.H.; Sun, S.N.; Wang, W.; Xu, Z.H.; Yu, S.J.; Jin, K.; Jin, Z.B. Effects of fluorescent protein tdTomato on mouse retina. Exp. Eye Res. 2024, 243, 109910. [Google Scholar] [CrossRef]
- Wang, D.; Zhong, Y.; Feng, B.; Qi, X.; Yan, T.; Liu, J.; Guo, S.; Wang, Y.; Liu, Z.; Cheng, D.; et al. The RUBY reporter enables efficient haploid identification in maize and tomato. Plant Biotechnol. J. 2023, 21, 1707–1715. [Google Scholar] [CrossRef]
- Ahmed, F.H.; Caputo, A.T.; French, N.G.; Peat, T.S.; Whitfield, J.; Warden, A.C.; Newman, J.; Scott, C. Over the rainbow: Structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed. Acta Crystallogr. D Struct. Biol. 2022, 78, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Casareto, B.E.; Yucharoen, M.; Dohra, H.; Suzuki, Y. Coexistence of nonfluorescent chromoproteins and fluorescent proteins in massive Porites spp. corals manifesting a pink pigmentation response. Front. Physiol. 2024, 15, 1339907. [Google Scholar] [CrossRef] [PubMed]
- Battad, J.M.; Wilmann, P.G.; Olsen, S.; Byres, E.; Smith, S.C.; Dove, S.G.; Turcic, K.N.; Devenish, R.J.; Rossjohn, J.; Prescott, M. A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins. J. Mol. Biol. 2007, 368, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Murakoshi, H.; Horiuchi, H.; Kosugi, T.; Onda, M.; Sato, A.; Koga, N.; Nabekura, J. ShadowR: A novel chromoprotein with reduced non-specific binding and improved expression in living cells. Sci. Rep. 2019, 9, 12072. [Google Scholar] [CrossRef] [PubMed]
- Tafoya-Ramirez, M.D.; Padilla-Vaca, F.; Ramirez-Saldana, A.P.; Mora-Garduno, J.D.; Rangel-Serrano, A.; Vargas-Maya, N.I.; Herrera-Gutierrez, L.J.; Franco, B. Replacing Standard Reporters from Molecular Cloning Plasmids with Chromoproteins for Positive Clone Selection. Molecules 2018, 23, 1328. [Google Scholar] [CrossRef]
- Tian, M.; Wei, J.; Lv, E.; Li, C.; Liu, G.; Sun, Y.; Yang, W.; Wang, Q.; Shen, C.; Zhang, C.; et al. Drug evaluation platform based on non-destructive and real-time in situ organoid fate state monitoring by graphene field-effect transistor. Chem. Eng. J. 2024, 498, 155355. [Google Scholar] [CrossRef]
- Wannier, T.M.; Mayo, S.L. The structure of a far-red fluorescent protein, AQ143, shows evidence in support of reported red-shifting chromophore interactions. Protein Sci. 2014, 23, 1148–1153. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, G.; Zha, C.; Su, J.; Cheng, F.; Tang, J.; Xu, X.; Li, J.; Wang, W.; Liu, Y. Colored Proteins Act as Biocolorants in Escherichia coli. Molecules 2025, 30, 432. https://doi.org/10.3390/molecules30030432
Sun G, Zha C, Su J, Cheng F, Tang J, Xu X, Li J, Wang W, Liu Y. Colored Proteins Act as Biocolorants in Escherichia coli. Molecules. 2025; 30(3):432. https://doi.org/10.3390/molecules30030432
Chicago/Turabian StyleSun, Geng, Chunmei Zha, Jingwen Su, Feng Cheng, Jian Tang, Xiuquan Xu, Jincai Li, Wenjian Wang, and Yu Liu. 2025. "Colored Proteins Act as Biocolorants in Escherichia coli" Molecules 30, no. 3: 432. https://doi.org/10.3390/molecules30030432
APA StyleSun, G., Zha, C., Su, J., Cheng, F., Tang, J., Xu, X., Li, J., Wang, W., & Liu, Y. (2025). Colored Proteins Act as Biocolorants in Escherichia coli. Molecules, 30(3), 432. https://doi.org/10.3390/molecules30030432





