CRISPR-Cas9 Gene Therapy: Non-Viral Delivery and Stimuli-Responsive Nanoformulations
Abstract
1. Introduction
2. CRISPR-Cas9 Gene Editing Mechanism

3. Non-Viral Delivery System of CRISPR-Cas9
3.1. Lipid-Based Nanoparticles
3.1.1. Structure and Characteristics of Lipid Nanoparticles (LNPs)
3.1.2. Advantages of LNPs in CRISPR-Cas9 Delivery
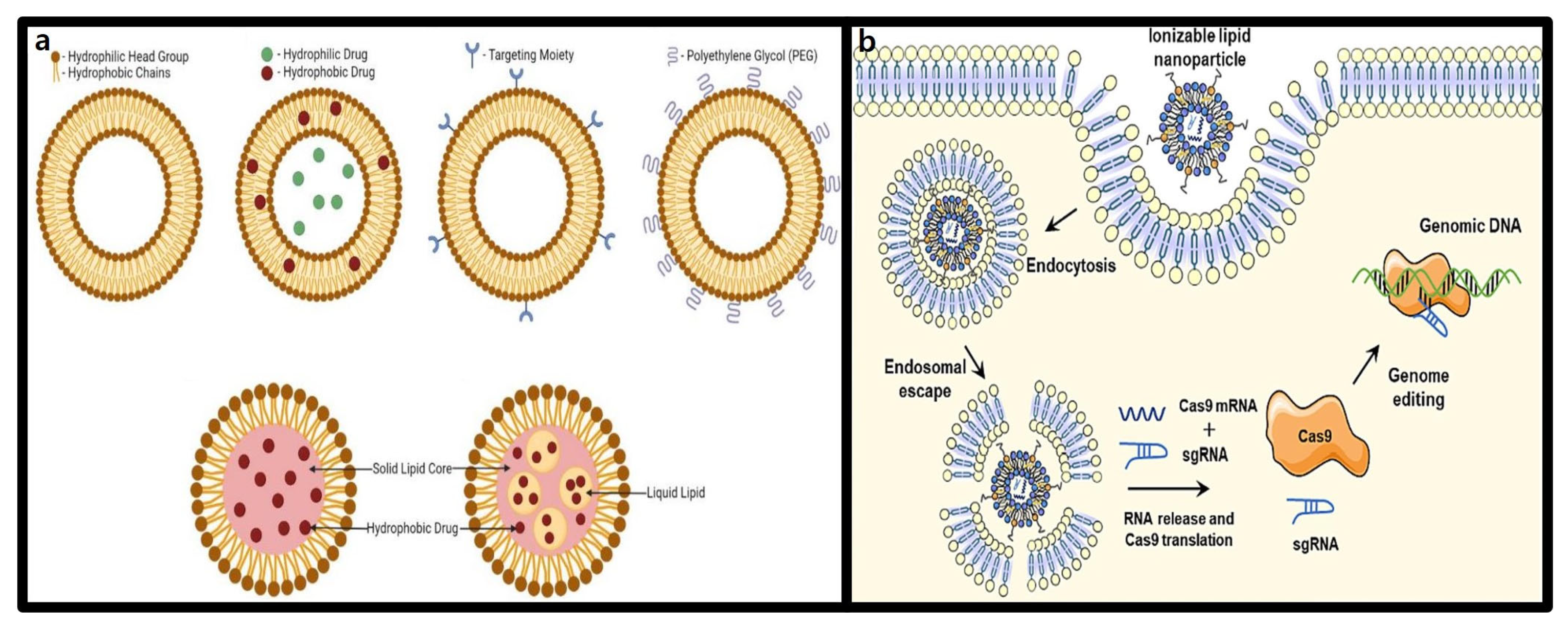
3.2. Extracellular Vesicles
3.2.1. Structure and Characteristics of Extracellular Vesicles (EVs)
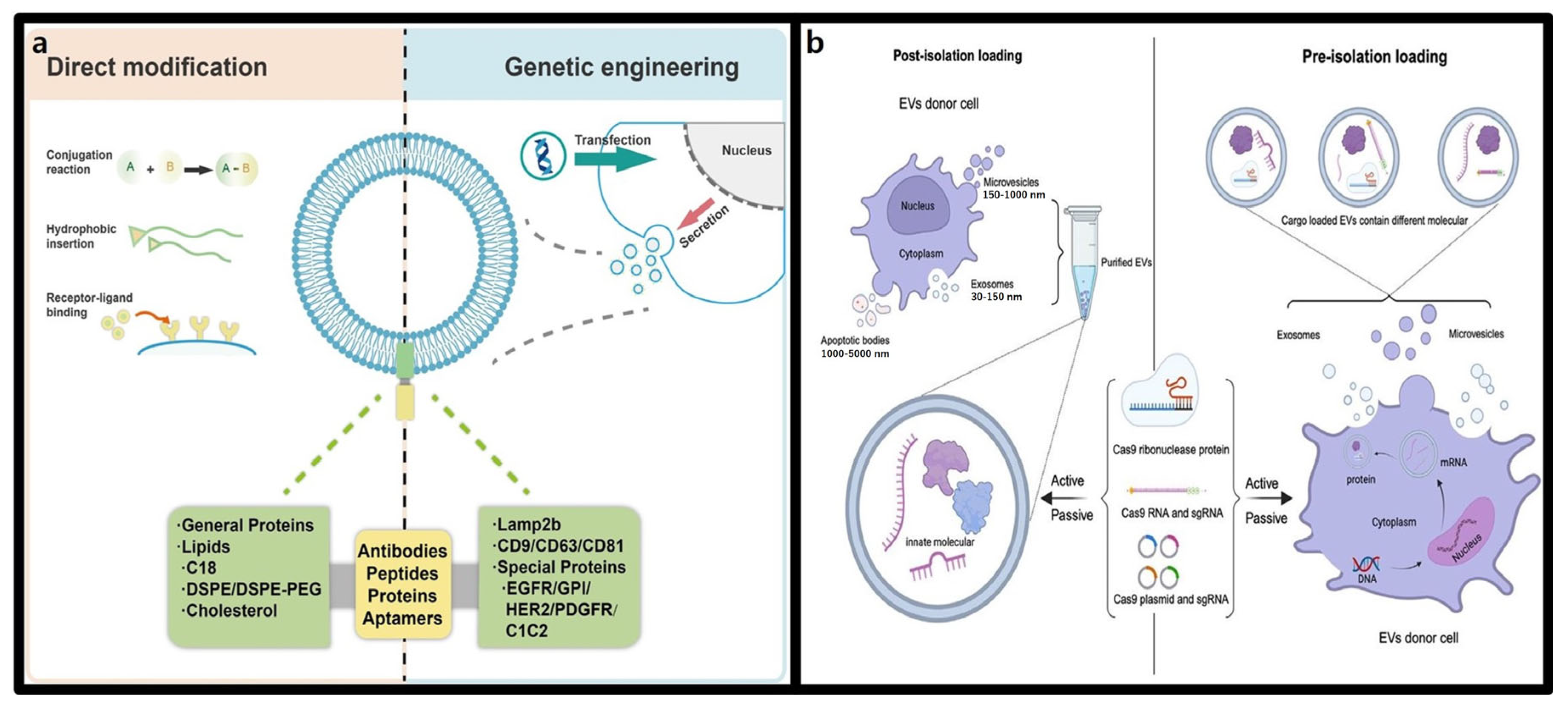
3.2.2. Advantages of Extracellular Vesicles in CRISPR-Cas9 Delivery
3.3. Polymer-Based Nanoparticles
3.3.1. Structure and Characteristics of Polymer-Based Nanoparticles
3.3.2. Advantages of Polymer-Based Nanoparticles in CRISPR-Cas9 Delivery
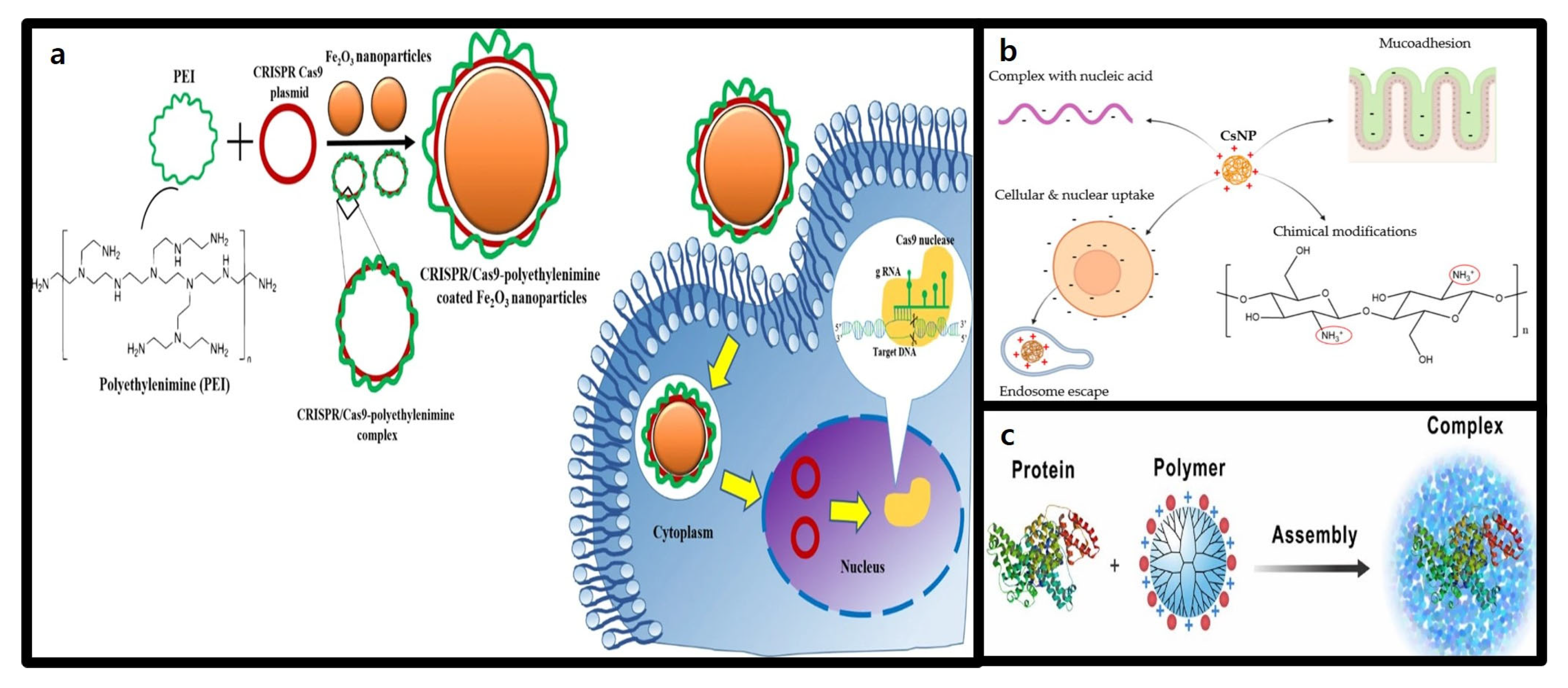
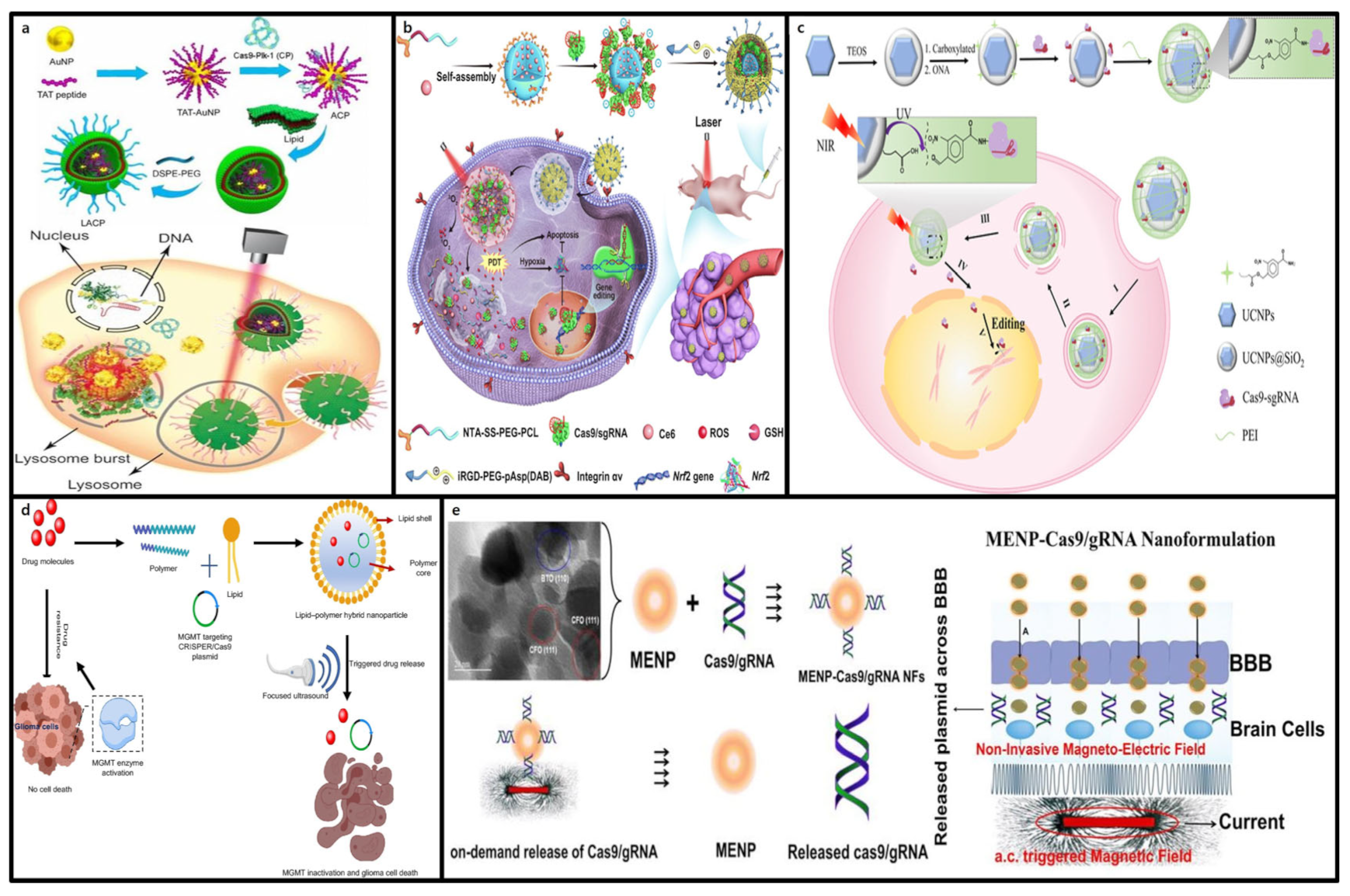
3.4. Gold Nanoparticles
3.4.1. Structure and Characteristics of Gold Nanoparticles (AuNPs)
3.4.2. Advantages of Gold Nanoparticles in CRISPR-Cas9 Delivery
3.5. Mesoporous Silica Nanoparticles (MSNs)
3.5.1. Structure and Characteristics of Mesoporous Silica Nanoparticles (MSNs)
3.5.2. Advantages of Mesoporous Silica Nanoparticles (MSNs) in CRISPR-Cas9 Delivery

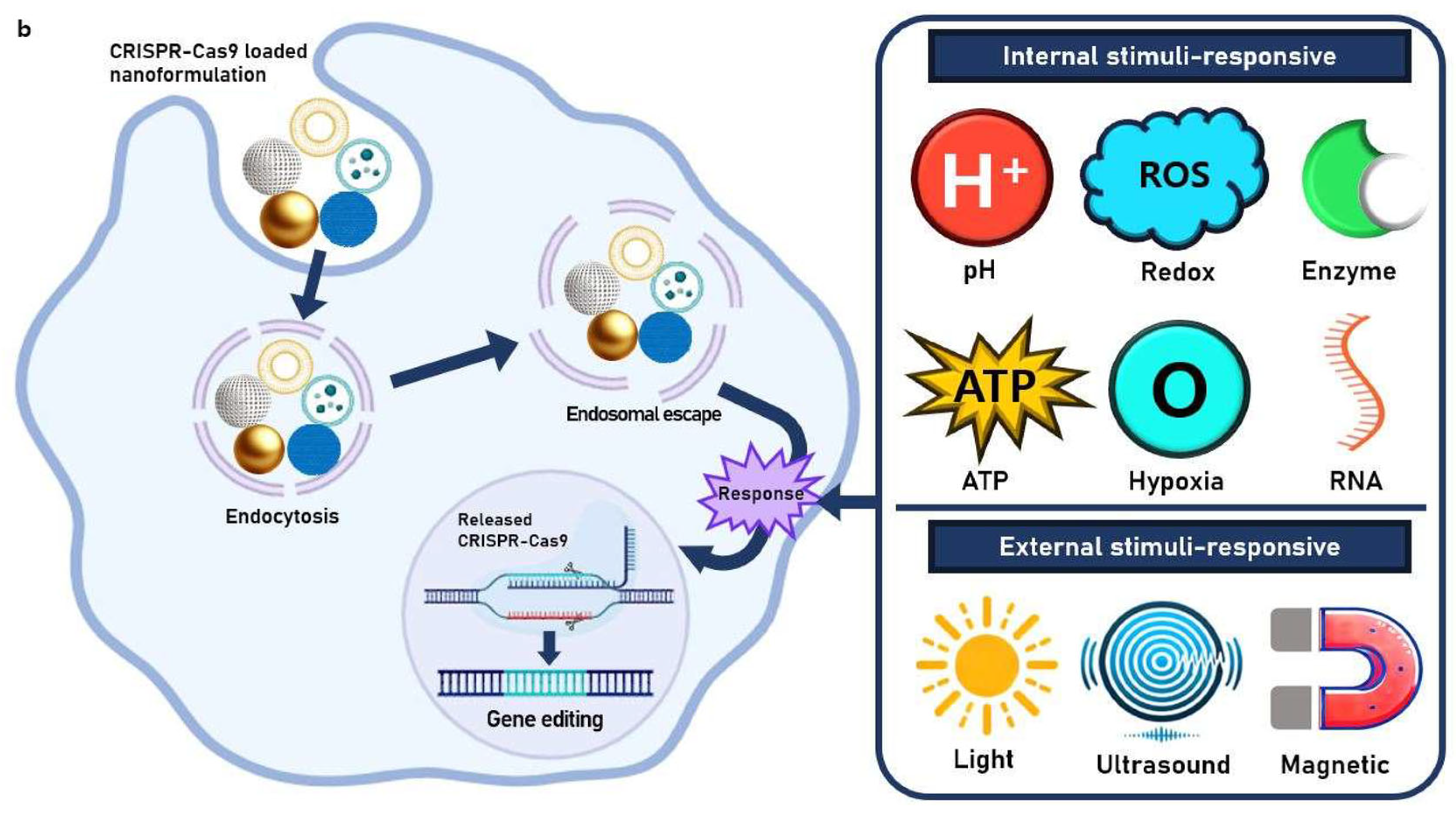
4. Stimuli-Responsive Nanoformulations
4.1. Internal Stimuli-Responsive Nanoformulations
4.1.1. pH-Responsive CRISPR-Cas9 Delivery
4.1.2. ATP-Responsive CRISPR-Cas9 Delivery
4.1.3. Redox-Responsive CRISPR-Cas9 Delivery
4.1.4. Hypoxia-Responsive CRISPR-Cas9 Delivery
4.1.5. Enzyme-Responsive CRISPR-Cas9 Delivery
4.1.6. RNA-Responsive CRISPR-Cas9 Delivery
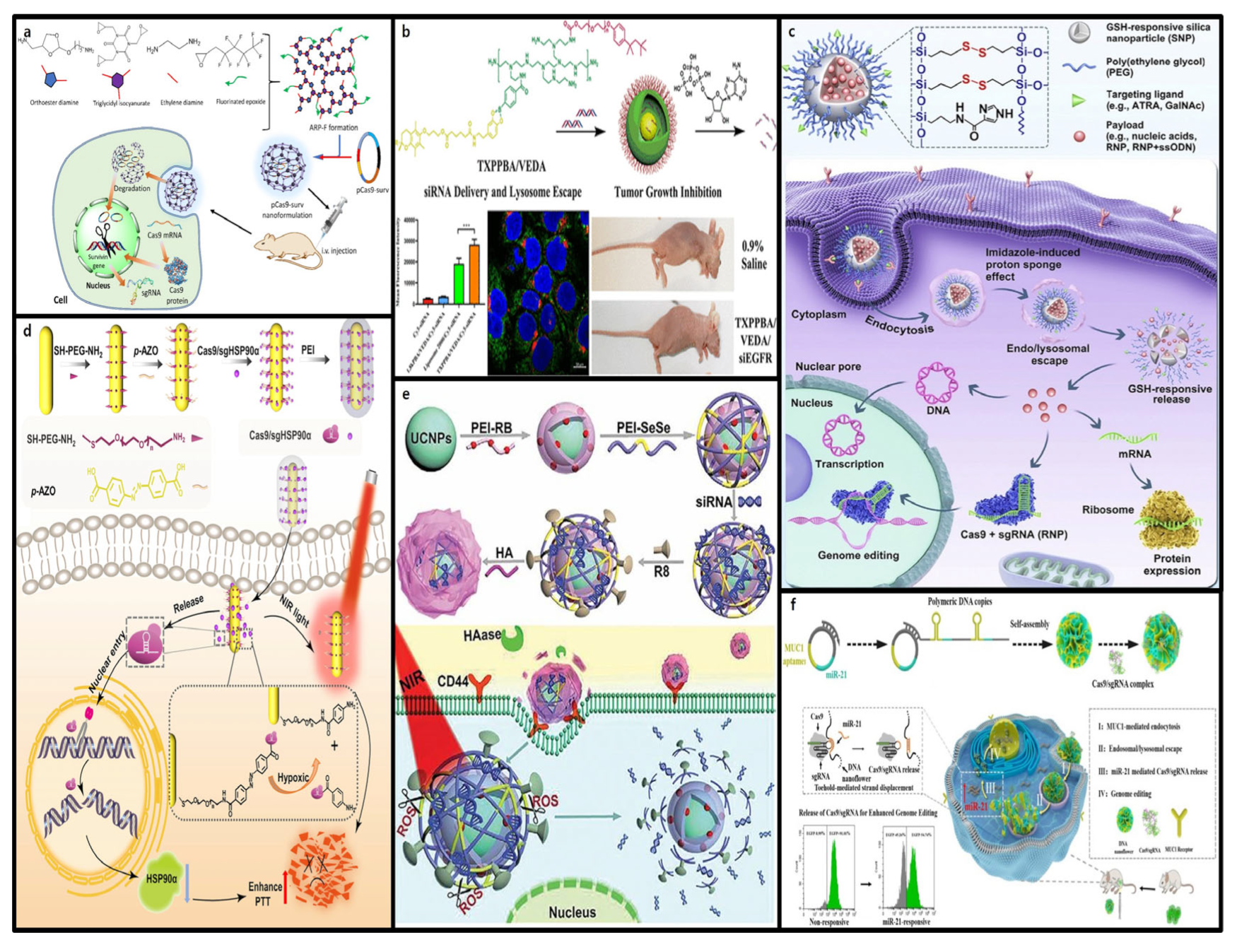
4.2. External Stimuli-Responsive Nanoformulations
4.2.1. Light-Responsive CRISPR-Cas9 Delivery
4.2.2. Ultrasound-Responsive CRISPR-Cas9 Delivery
4.2.3. Magnetic-Responsive CRISPR-Cas9 Delivery
5. Perspective: Towards Clinical Application of Advanced CRISPR-Cas9 Delivery Systems
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sayed, N.; Allawadhi, P.; Khurana, A.; Singh, V.; Navik, U.; Pasumarthi, S.K.; Khurana, I.; Banothu, A.K.; Weiskirchen, R.; Bharani, K.K. Gene therapy: Comprehensive overview and therapeutic applications. Life Sci. 2022, 294, 120375. [Google Scholar] [CrossRef]
- Lu, X.-J.; Xue, H.-Y.; Ke, Z.-P.; Chen, J.-L.; Ji, L.-J. CRISPR-Cas9: A new and promising player in gene therapy. J. Med. Genet. 2015, 52, 289–296. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.R.; Markusic, D.M.; Biswas, M.; High, K.A.; Herzog, R.W. Clinical development of gene therapy: Results and lessons from recent successes. Mol. Ther. Methods Clin. Dev. 2016, 3, 16034. [Google Scholar] [CrossRef] [PubMed]
- Cring, M.R.; Sheffield, V.C. Gene therapy and gene correction: Targets, progress, challenges for treating human diseases. Gene Ther. 2022, 29, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M. The genome editing revolution: Review. J. Genet. Eng. Biotechnol. 2020, 18, 68. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, H.; Fan, X.; Zhang, Y.; Zhang, M.; Wang, Y.; Xie, Z.; Bai, M.; Yin, Q.; Liang, D.; et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015, 25, 67–79. [Google Scholar] [CrossRef]
- Tong, L.W.; Hu, Y.S.; Yu, S.J.; Li, C.L.; Shao, J.W. Current application and future perspective of CRISPR/cas9 gene editing system mediated immune checkpoint for liver cancer treatment. Nanotechnology 2024, 35, ad5f33. [Google Scholar] [CrossRef]
- Alhakamy, N.A.; Curiel, D.T.; Berkland, C.J. The era of gene therapy: From preclinical development to clinical application. Drug Discov. Today 2021, 26, 1602–1619. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, W.-J.; Kim, C.H.; Oh, Y.; Gwon, L.W.; Lee, H.; Song, W.; Hur, J.K.; Lim, K.-S.; Jeong, K.J.; et al. Highly specific chimeric DNA-RNA-guided genome editing with enhanced CRISPR-Cas12a system. Mol. Ther. -Nucleic Acids 2022, 28, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Luther, D.C.; Lee, Y.W.; Nagaraj, H.; Scaletti, F.; Rotello, V.M. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: Advances and challenges. Expert. Opin. Drug Deliv. 2018, 15, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release 2017, 266, 17–26. [Google Scholar] [CrossRef]
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef]
- Xu, X.; Wan, T.; Xin, H.; Li, D.; Pan, H.; Wu, J.; Ping, Y. Delivery of CRISPR/Cas9 for therapeutic genome editing. J. Gene Med. 2019, 21, e3107. [Google Scholar] [CrossRef] [PubMed]
- Sioson, V.A.; Kim, M.; Joo, J. Challenges in delivery systems for CRISPR-based genome editing and opportunities of nanomedicine. Biomed. Eng. Lett. 2021, 11, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Singh, D. Revolutionizing Lung Cancer Treatment: Innovative CRISPR-Cas9 Delivery Strategies. AAPS PharmSciTech 2024, 25, 129. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Huang, K.; Zapata, D.; Tang, Y.; Teng, Y.; Li, Y. In vivo delivery of CRISPR-Cas9 genome editing components for therapeutic applications. Biomaterials 2022, 291, 121876. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Song, C.-Q.; Dorkin, J.R.; Zhu, L.J.; Li, Y.; Wu, Q.; Park, A.; Yang, J.; Suresh, S.; Bizhanova, A.; et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016, 34, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, X.; Zhang, Y.; Wang, H.; Ying, H.; Liu, M.; Li, D.; O Lui, K.; Ding, Q. A Self-restricted CRISPR System to Reduce Off-target Effects. Mol. Ther. 2016, 24, 1508–1510. [Google Scholar] [CrossRef]
- Ehrke-Schulz, E.; Schiwon, M.; Leitner, T.; Dávid, S.; Bergmann, T.; Liu, J.; Ehrhardt, A. CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci. Rep. 2017, 7, 17113. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Glass, Z.A.; Xu, Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017, 24, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Zu, H.; Gao, D. Non-viral Vectors in Gene Therapy: Recent Development, Challenges, Prospects. AAPS J. 2021, 23, 78. [Google Scholar] [CrossRef] [PubMed]
- Timin, A.S.; Muslimov, A.R.; Lepik, K.V.; Epifanovskaya, O.S.; Shakirova, A.I.; Mock, U.; Riecken, K.; Okilova, M.V.; Sergeev, V.S.; Afanasyev, B.V.; et al. Efficient gene editing via non-viral delivery of CRISPR–Cas9 system using polymeric and hybrid microcarriers. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Upponi, J.R.; Torchilin, V. Design of multifunctional non-viral gene vectors to overcome physiological barriers: Dilemmas and strategies. Int. J. Pharm. 2012, 427, 3–20. [Google Scholar] [CrossRef] [PubMed]
- English, M.A.; Soenksen, L.R.; Gayet, R.V.; de Puig, H.; Angenent-Mari, N.M.; Mao, A.S.; Nguyen, P.Q.; Collins, J.J. Programmable CRISPR-responsive smart materials. Science 2019, 365, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Teng, L.; Gao, L.; Su, T.; Fu, L.; Qiu, Z.; Bi, Y. Advances in multiple stimuli-responsive drug-delivery systems for cancer therapy. Int. J. Nanomed. 2021, 16, 1525–1551. [Google Scholar] [CrossRef]
- Hasanzadeh, A.; Noori, H.; Jahandideh, A.; Moghaddam, N.H.; Mousavi, S.M.K.; Nourizadeh, H.; Saeedi, S.; Karimi, M.; Hamblin, M.R. Smart Strategies for Precise Delivery of CRISPR/Cas9 in Genome Editing. ACS Appl. Bio Mater. 2022, 5, 413–437. [Google Scholar] [CrossRef]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef]
- Fonfara, I.; Richter, H.; Bratovič, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Mitrofanov, A.; Ziemann, M.; Alkhnbashi, O.S.; Hess, W.R.; Backofen, R. CRISPRtracrRNA: Robust approach for CRISPR tracrRNA detection. Bioinformatics 2022, 38, ii42–ii48. [Google Scholar] [CrossRef]
- Kim, H.; Lee, W.J.; Oh, Y.; Kang, S.H.; Hur, J.K.; Lee, H. Enhancement of target specificity of CRISPR–Cas12a by using a chimeric DNA–RNA guide. Nucleic Acids Res. 2020, 48, 8601–8616. [Google Scholar] [CrossRef]
- Gorski, S.A.; Vogel, J.; Doudna, J.A. RNA-based recognition and targeting: Sowing the seeds of specificity. Nat. Rev. Mol. Cell Biol. 2017, 18, 215–228. [Google Scholar] [CrossRef]
- Cui, Y.; Xu, J.; Cheng, M.; Liao, X.; Peng, S. Review of CRISPR/Cas9 sgRNA Design Tools. Interdiscip. Sci. 2018, 10, 455–465. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Huai, C.; Li, G.; Yao, R.; Zhang, Y.; Cao, M.; Kong, L.; Jia, C.; Yuan, H.; Chen, H.; Lu, D.; et al. Structural insights into DNA cleavage activation of CRISPR-Cas9 system. Nat. Commun. 2017, 8, 1375. [Google Scholar] [CrossRef] [PubMed]
- Han, H.A.; Pang, J.K.S.; Soh, B.S. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J. Mol. Med. 2020, 98, 615–632. [Google Scholar] [CrossRef]
- Kang, S.; Jeon, S.; Kim, S.; Chang, Y.K.; Kim, Y.C. Development of a pVEC peptide-based ribonucleoprotein (RNP) delivery system for genome editing using CRISPR/Cas9 in Chlamydomonas reinhardtii. Sci. Rep. 2020, 10, 22158. [Google Scholar] [CrossRef]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Corrigendum: Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2016, 34, 210. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Chu, V.T.; Weber, T.; Wefers, B.; Wurst, W.; Sander, S.; Rajewsky, K.; Kühn, R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015, 33, 543–548. [Google Scholar] [CrossRef]
- Jayavaradhan, R.; Pillis, D.M.; Goodman, M.; Zhang, F.; Zhang, Y.; Andreassen, P.R.; Malik, P. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed]
- Fellmann, C.; Gowen, B.G.; Lin, C.; Doudna, J.A.; Corn, J.E. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017, 16, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wagner, E.; Lächelt, U. Non-viral delivery of the CRISPR/Cas system: DNA. Biomater. Sci. 2022, 10, 1166–1192. [Google Scholar] [CrossRef]
- Gu, Y.; Gao, J.; Cao, M.; Dong, C.; Lian, J.; Huang, L.; Cai, J.; Xu, Z. Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris. World J. Microbiol. Biotechnol. 2019, 35, 79. [Google Scholar] [CrossRef] [PubMed]
- Lauritsen, I.; Porse, A.; Sommer, M.O.A.; Nørholm, H.M.H. A versatile one-step CRISPR-Cas9 based approach to plasmid-curing. Microb. Cell Fact. 2017, 16, 135. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, J.N.; Geng, K.; Sommerauer, C.; Atanasoai, I.; Yin, X.; Kutter, C. Successful delivery of large-size CRISPR/Cas9 vectors in hard-to-transfect human cells using small plasmids. Commun. Biol. 2020, 3, 319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Zhang, Y.; Yin, H. Genome Editing with mRNA Encoding ZFN, TALEN, Cas9. Mol. Ther. 2019, 27, 735–746. [Google Scholar] [CrossRef]
- Low, B.E.; Kutny, M.; Simple, M.V.W. Efficient CRISPR-Cas9-Mediated Gene Editing in Mice: Strategies and Methods. Methods Mol. Biol. 2016, 1438, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Eftekhari, Z.; Zohrabi, H.; Oghalaie, A.; Ebrahimi, T.; Shariati, F.S.; Behdani, M.; Kazemi-Lomedasht, F. Advancements and challenges in mRNA and ribonucleoprotein-based therapies: From delivery systems to clinical applications. Mol. Ther. Nucleic Acids 2024, 35, 102313. [Google Scholar] [CrossRef]
- Wei, T.; Cheng, Q.; Min, Y.L.; Olson, E.N.; Siegwart, D.J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 2020, 11, 3232. [Google Scholar] [CrossRef]
- Sahel, D.K.; Vora, L.K.; Saraswat, A.; Sharma, S.; Monpara, J.; D’Souza, A.A.; Mishra, D.; Tryphena, K.P.; Kawakita, S.; Khan, S.; et al. CRISPR/Cas9 Genome Editing for Tissue-Specific In Vivo Targeting: Nanomaterials and Translational Perspective. Adv. Sci. 2023, 10, e2305072. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics 2021, 11, 614–648. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, F.; Begum, A.A.; Dai, C.C.; Toth, I.; Moyle, M. Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing. Drug Deliv. Transl. Res. 2023, 13, 1500–1519. [Google Scholar] [CrossRef]
- van Kampen, S.J.; van Rooij, E. CRISPR Craze to Transform Cardiac Biology. Trends Mol. Med. 2019, 25, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, Y.; Hu, J.; Peng, X.; Liu, Z. CRISPR/Cas9 systems: Delivery technologies and biomedical applications. Asian J. Pharm. Sci. 2023, 18, 100854. [Google Scholar] [CrossRef]
- Kang, S.-H.; Lee, W.-J.; An, J.-H.; Lee, J.-H.; Kim, Y.-H.; Kim, H.; Oh, Y.; Park, Y.-H.; Jin, Y.B.; Jun, B.-H.; et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nat. Commun. 2020, 11, 3596. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y. Application of the CRISPR/Cas9 system to drug resistance in breast cancer. Adv. Sci. 2018, 5, 1700964. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Y.; Yang, J.; Li, W.; Zhang, M.; Wang, Q.; Zhang, L.; Wei, G.; Tian, Y.; Zhao, K.; et al. Non-viral, specifically targeted CAR-T cells achieve high safety and efficacy in B-NHL. Nature 2022, 609, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Alsaiari, S.K.; Eshaghi, B.; Du, B.; Kanelli, M.; Li, G.; Wu, X.; Zhang, L.; Chaddah, M.; Lau, A.; Yang, X.; et al. CRISPR–Cas9 delivery strategies for the modulation of immune and non-immune cells. Nat. Rev. Mater. 2025, 10, 44–61. [Google Scholar] [CrossRef]
- Rohner, E.; Yang, R.; Foo, K.S.; Goedel, A.; Chien, K.R. Unlocking the promise of mRNA therapeutics. Nat. Biotechnol. 2022, 40, 1586–1600. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef]
- Jain, V.; Kumar, H.; Anod, H.V.; Chand, P.; Gupta, N.V.; Dey, S.; Kesharwani, S.S. A review of nanotechnology-based approaches for breast cancer and triple-negative breast cancer. J. Control. Release 2020, 326, 628–647. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.J.; Kulkarni, J.A.; van der Meel, R.; Cullis, R.; Vader, P.; Schiffelers, R.M. State-of-the-art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods 2018, 2, 1700375. [Google Scholar] [CrossRef]
- Kenjo, E.; Hozumi, H.; Makita, Y.; Iwabuchi, K.A.; Fujimoto, N.; Matsumoto, S.; Kimura, M.; Amano, Y.; Ifuku, M.; Naoe, Y.; et al. Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice. Nat. Commun. 2021, 12, 7101. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Li, X. Liposomal delivery of CRISPR/Cas9. Cancer Gene Ther. 2020, 27, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Magar, K.T.; Boafo, G.F.; Li, X.; Chen, Z.; He, W. Liposome-based delivery of biological drugs. Chin. Chem. Lett. 2022, 33, 587–596. [Google Scholar] [CrossRef]
- Meng, N.; Grimm, D. Membrane-destabilizing ionizable phospholipids: Novel components for organ-selective mRNA delivery and CRISPR–Cas gene editing. Signal Transduct. Target. Ther. 2021, 6, 206. [Google Scholar] [CrossRef] [PubMed]
- Leong, E.W.X.; Ge, R. Lipid Nanoparticles as Delivery Vehicles for Inhaled Therapeutics. Biomedicines 2022, 10, 2179. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, D.; Gutkin, A.; Kedmi, R.; Ramishetti, S.; Veiga, N.; Jacobi, A.M.; Schubert, M.S.; Friedmann-Morvinski, D.; Cohen, Z.R.; Behlke, M.A.; et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020, 6, eabc9450. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, C.H.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, H.; Ding, F.; Gao, X.; Zhu, Z.; Yang, C. Development of ionizable lipid nanoparticles and a lyophilized formulation for potent CRISPR-Cas9 delivery and genome editing. Int. J. Pharm. 2024, 652, 123845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441. [Google Scholar] [CrossRef]
- Kim, M.; Jeong, M.; Hur, S.; Cho, Y.; Park, J.; Jung, H.; Seo, Y.; Woo, H.A.; Nam, K.T.; Lee, K.; et al. Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci. Adv. 2021, 7, eabf4398. [Google Scholar] [CrossRef]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’connell, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Prakash, G.; Shokr, A.; Willemen, N.; Bashir, S.M.; Shin, S.R.; Hassan, S. Microfluidic fabrication of lipid nanoparticles for the delivery of nucleic acids. Adv. Drug Deliv. Rev. 2022, 184, 114197. [Google Scholar] [CrossRef] [PubMed]
- Majeau, N.; Fortin-Archambault, A.; Gérard, C.; Rousseau, J.; Yaméogo, J.; Tremblay, J. Serum extracellular vesicles for delivery of CRISPR-CAS9 ribonucleoproteins to modify the dystrophin gene. Mol. Ther. 2022, 30, 2429–2442. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef]
- Chen, L.; Hong, W.; Ren, W.; Xu, T.; Qian, Z.; He, Z. Recent progress in targeted delivery vectors based on biomimetic nanoparticles. Signal Transduct. Target. Ther. 2021, 6, 225. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, L.; Zeng, X.; Schwarz, H.; Nanda, H.S.; Peng, X.; Zhou, Y. Exosomes, a New Star for Targeted Delivery. Front. Cell Dev. Biol. 2021, 9, 751079. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Han, Y.; An, Y.; Ding, Y.; He, C.; Wang, X.; Tang, Q. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials 2018, 178, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Duan, J.; Liu, R.; Du, Y.; Luo, Q.; Cui, Y.; Su, Z.; Xu, J.; Xie, Y.; Lu, W. Engineered targeting tLyp-1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J. Pharm. Sci. 2020, 15, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Parada, N.; Romero-Trujillo, A.; Georges, N.; Alcayaga-Miranda, F. Camouflage strategies for therapeutic exosomes evasion from phagocytosis. J. Adv. Res. 2021, 31, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, S.; Liu, L.; Dang, P.; Liu, Y.; Sun, Z.; Qiao, B.; Wang, C. Engineered exosomes from different sources for cancer-targeted therapy. Signal Transduct. Target. Ther. 2023, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Godbout, K.; Lamothe, G.; Tremblay, J. CRISPR-Cas9 delivery strategies with engineered extracellular vesicles. Mol. Ther. Nucleic Acids 2023, 34, 102040. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control. Release 2015, 205, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Sil, S.; Dagur, R.S.; Liao, K.; Peeples, E.S.; Hu, G.; Periyasamy, P.; Buch, S. Strategies for the use of extracellular vesicles for the delivery of therapeutics. J. Neuroimmune Pharmacol. 2020, 15, 422–442. [Google Scholar] [CrossRef]
- Lains, D.K.; Kadunc, L.; Keber, M.M.E.; Bratkovic, I.H.; Romih, R.; Jerala, R. Delivery of an artificial transcription regulator dCas9-VPR by extracellular vesicles for therapeutic gene activation. ACS Synth. Biol. 2018, 7, 2715–2725. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Liang, Y.; Xu, X.; Xu, L.; Iqbal, Z.; Ouyang, K.; Zhang, H.; Wen, C.; Duan, L.; Xia, J. Chondrocyte-specific genomic editing enabled by hybrid exosomes for osteoarthritis treatment. Theranostics 2022, 12, 4866. [Google Scholar] [CrossRef]
- Fortuni, B.; Inose, T.; Ricci, M.; Fujita, Y.; Van Zundert, I.; Masuhara, A.; Fron, E.; Mizuno, H.; Latterini, L.; Rocha, S.; et al. Polymeric engineering of nanoparticles for highly efficient multifunctional drug delivery systems. Sci. Rep. 2019, 9, 2666. [Google Scholar] [CrossRef]
- Sung, Y.K.; Kim, S.W. Recent advances in polymeric drug delivery systems. Biomater. Res. 2020, 24, 12. [Google Scholar] [CrossRef]
- Piotrowski-Daspit, A.S.; Kauffman, A.C.; Bracaglia, L.G.; Saltzman, W.M. Polymeric vehicles for nucleic acid delivery. Adv. Drug Deliv. Rev. 2020, 156, 119–132. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, M.S.; Susnik, E.; Drasler, B.; Taladriz-Blanco, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem. Soc. Rev. 2021, 50, 5397–5434. [Google Scholar] [CrossRef]
- Zhao, H.; Lin, Z.Y.; Yildirimer, L.; Dhinakar, A.; Zhao, X.; Wu, J. Polymer-based nanoparticles for protein delivery: Design, strategies and applications. J. Mater. Chem. B 2016, 4, 4060–4071. [Google Scholar] [CrossRef] [PubMed]
- Ryu, N.; Kim, M.-A.; Park, D.; Lee, B.; Kim, Y.-R.; Kim, K.-H.; Baek, J.-I.; Kim, W.J.; Lee, K.-Y.; Kim, U.-K. Effective PEI-mediated delivery of CRISPR-Cas9 complex for targeted gene therapy. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 2095–2102. [Google Scholar] [CrossRef]
- Ahern, J.O.; Lara-Sáez, I.; Zhou, D.; Murillas, R.; Bonafont, J.; Mencía, Á.; García, M.; Manzanares, D.; Lynch, J.; Foley, R.; et al. Non-viral delivery of CRISPR–Cas9 complexes for targeted gene editing via a polymer delivery system. Gene Ther. 2022, 29, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Caprifico, A.E.; Foot, J.S.; Polycarpou, E.; Calabrese, G. Advances in Chitosan-Based CRISPR/Cas9 Delivery Systems. Pharm. 2022, 14, 1840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wan, T.; Chen, Y.; Chen, Y.; Sun, H.; Cao, T.; Songyang, Z.; Tang, G.; Wu, C.; Ping, Y.; et al. Cationic polymer-mediated CRISPR/Cas9 plasmid delivery for genome editing. Macromol. Rapid Commun. 2019, 40, 1800068. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-Y.; He, X.-Y.; Xu, C.; Xu, L.; Ai, S.-L.; Cheng, S.-X.; Zhuo, R.-X. A dual-targeting delivery system for effective genome editing and in situ detecting related protein expression in edited cells. Biomacromolecules 2018, 19, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wan, T.; Wang, H.; Zhang, S.; Ping, Y.; Cheng, Y. A boronic acid-rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci Adv. 2019, 5, eaaw8922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, G.; Yu, X.; Wei, T.; Farbiak, L.; Johnson, L.T.; Taylor, A.M.; Xu, J.; Hong, Y.; Zhu, H.; et al. Enhancing CRISPR/Cas gene editing through modulating cellular mechanical properties for cancer therapy. Nat. Nanotechnol. 2022, 17, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [PubMed]
- Srivastav, A.; Gupta, K.; Chakraborty, D.; Dandekar, P.; Jain, R. Efficiency of chitosan-coated PLGA nanocarriers for cellular delivery of siRNA and CRISPR/Cas9 complex. J. Pharm. Innov. 2020, 17, 180–193. [Google Scholar] [CrossRef]
- Rohiwal, S.S.; Dvorakova, N.; Klima, J.; Vaskovicova, M.; Senigl, F.; Slouf, M.; Pavlova, E.; Stepanek, P.; Babuka, D.; Benes, H.; et al. Polyethylenimine based magnetic nanoparticles mediated non-viral CRISPR/Cas9 system for genome editing. Sci. Rep. 2020, 10, 4619. [Google Scholar] [CrossRef]
- Chen, Z.; Li, J.; Li, T.; Fan, T.; Meng, C.; Li, C. A CRISPR/Cas12a-empowered surface plasmon resonance platform for rapid and specific diagnosis of the Omicron variant of SARS-CoV-2. Natl. Sci. Rev. 2022, 9, nwac104. [Google Scholar] [CrossRef] [PubMed]
- Heo, D.N.; Yang, D.H.; Moon, H.-J.; Lee, J.B.; Bae, M.S.; Lee, S.C.; Lee, W.J.; Sun, I.-C.; Kwon, I.K. Gold nanoparticles surface-functionalized with paclitaxel drug and biotin receptor as theranostic agents for cancer therapy. Biomaterials 2012, 33, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Connor, D.M.; Broome, A.-M. Gold nanoparticles for the delivery of cancer therapeutics. Adv. Cancer Res. 2018, 139, 163–184. [Google Scholar] [PubMed]
- Mout, R.; Ray, M.; Tonga, G.Y.; Lee, Y.-W.; Tay, T.; Sasaki, K.; Rotello, V.M. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 2017, 11, 2452–2458. [Google Scholar] [CrossRef]
- Yanik, M.; Müller, B.; Song, F.; Gall, J.; Wagner, F.; Wende, W.; Lorenz, B.; Stieger, K. In vivo genome editing as a potential treatment strategy for inherited retinal dystrophies. Progress. Retin. Eye Res. 2017, 56, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, K.; Panda, S.; Gonzales-Rojas, R.; Chong, A.; Bugay, V.; Park, H.M.; Brenner, R.; Murthy, N.; Lee, H.Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, N.; Dykman, L. Biodistribution and toxicity of engineered gold nanoparticles: A review of in vitro and in vivo studies. Chem. Soc. Rev. 2011, 40, 1647–1671. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Q.; Han, N.; Bai, L.; Li, J.; Liu, J.; Che, E.; Hu, L.; Zhang, Q.; Jiang, T.; et al. Mesoporous silica nanoparticles in drug delivery and biomedical applications. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 313–327. [Google Scholar] [CrossRef]
- Xu, X.; Koivisto, O.; Liu, C.; Zhou, J.; Miihkinen, M.; Jacquemet, G.; Wang, D.; Rosenholm, J.M.; Shu, Y.; Zhang, H. Effective delivery of the CRISPR/Cas9 system enabled by functionalized mesoporous silica nanoparticles for GFP-tagged paxillin knock-in. Adv. Ther. 2021, 4, 2000072. [Google Scholar] [CrossRef]
- Tarn, D.; Ashley, C.E.; Xue, M.; Carnes, E.C.; Zink, J.I.; Brinker, C.J. Mesoporous silica nanoparticle nanocarriers: Biofunctionality and biocompatibility. Acc. Chem. Res. 2013, 46, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Kovochich, M.; Liong, M.; Meng, H.; Kabehie, S.; George, S.; Zink, J.I.; Nel, A.E. Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano 2009, 3, 3273–3286. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Wang, H.; Ducheyne, P. Polymer-coated mesoporous silica nanoparticles for the controlled release of macromolecules. Acta Biomater. 2012, 8, 3429–3435. [Google Scholar] [CrossRef] [PubMed]
- Han, D.H.; Na, H.-K.; Choi, W.H.; Lee, J.H.; Kim, Y.K.; Won, C.; Lee, S.-H.; Kim, K.P.; Kuret, J.; Min, D.-H.; et al. Direct cellular delivery of human proteasomes to delay tau aggregation. Nat. Commun. 2014, 5, 5633. [Google Scholar] [CrossRef]
- Na, H.; Kim, M.; Park, K.; Ryoo, S.; Lee, K.E.; Jeon, H.; Ryoo, R.; Hyeon, C.; Min, D. Efficient functional delivery of siRNA using mesoporous silica nanoparticles with ultralarge pores. Small 2012, 8, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Encinas, N.; Angulo, M.; Astorga, C.; Colilla, M.; Izquierdo-Barba, I.; Vallet-Regí, M. Mixed-charge pseudo-zwitterionic mesoporous silica nanoparticles with low-fouling and reduced cell uptake properties. Acta Biomater. 2019, 84, 317–327. [Google Scholar] [CrossRef]
- Rosenholm, J.M.; Sahlgren, C.; Lindén, M. Towards multifunctional targeted drug delivery systems using mesoporous silica nanoparticles–opportunities & challenges. Nanoscale 2010, 2, 1870–1883. [Google Scholar] [PubMed]
- Yang, Y.; Zhao, W.; Tan, W.; Lai, Z.; Fang, D.; Jiang, L.; Zuo, C.; Yang, N.; Lai, Y. An Efficient Cell-Targeting Drug Delivery System Based on Aptamer-Modified Mesoporous Silica Nanoparticles. Nanoscale Res. Lett. 2019, 14, 390. [Google Scholar] [CrossRef]
- García-Fernández, A.; Vivo-Llorca, G.; Sancho, M.; García-Jareño, A.B.; Ramírez-Jiménez, L.; Barber-Cano, E.; Murguía, J.R.; Orzáez, M.; Sancenón, F.; Martínez-Máñez, R. Nanodevices for the efficient codelivery of CRISPR-Cas9 editing machinery and an entrapped cargo: A proposal for dual anti-inflammatory therapy. Pharmaceutics 2022, 14, 1495. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, N.; Kim, T.; Kim, J.; Hyeon, T. Multifunctional mesoporous silica nanocomposite nanoparticles for theranostic applications. Acc. Chem. Res. 2011, 44, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhong, Q.; Wang, Y.; Hu, P.; Zhong, W.; Huang, C.B. Chemically engineered mesoporous silica nanoparticles-based intelligent delivery systems for theranostic applications in multiple cancerous/non-cancerous diseases. Coord. Chem. Rev. 2022, 452, 214309. [Google Scholar] [CrossRef]
- Lérida-Viso, A.; Estepa-Fernández, A.; García-Fernández, A.; Martí-Centelles, V.; Martínez-Máñez, R. Biosafety of mesoporous silica nanoparticles; towards clinical translation. Adv. Drug Deliv. Rev. 2023, 201, 115049. [Google Scholar] [CrossRef] [PubMed]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef]
- Mashel, T.V.; Tarakanchikova, Y.V.; Muslimov, A.R.; Zyuzin, M.V.; Timin, A.S.; Lepik, K.V.; Fehse, B. Overcoming the delivery problem for therapeutic genome editing: Current status and perspective of non-viral methods. Biomaterials 2020, 258, 120282. [Google Scholar] [CrossRef]
- Dogbey, D.M.; Torres, V.E.S.; Fajemisin, E.; Mpondo, L.; Ngwenya, T.; Akinrinmade, O.A.; Perriman, A.W.; Barth, S. Technological advances in the use of viral and non-viral vectors for delivering genetic and non-genetic cargos for cancer therapy. Drug Deliv. Transl. Res. 2023, 13, 2719–2738. [Google Scholar] [CrossRef]
- Zhuo, C.; Zhang, J.; Lee, J.-H.; Jiao, J.; Cheng, D.; Liu, L.; Kim, H.-W.; Tao, Y.; Li, M. Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduct. Target. Ther. 2021, 6, 238. [Google Scholar] [CrossRef]
- Zhou, Z.; Vázquez-González, M.; Willner, I. Stimuli-responsive metal–organic framework nanoparticles for controlled drug delivery and medical applications. Chem. Soc. Rev. 2021, 50, 4541–4563. [Google Scholar] [CrossRef] [PubMed]
- Mura, S.; Nicolas, J.; Couvreur, P. Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 2013, 12, 991–1003. [Google Scholar] [CrossRef]
- Zhang, P.; Gao, D.; An, K.; Shen, Q.; Wang, C.; Zhang, Y.; Pan, X.; Chen, X.; Lyv, Y.; Cui, C.; et al. A programmable polymer library that enables the construction of stimuli-responsive nanocarriers containing logic gates. Nat. Chem. 2020, 12, 381–390. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Delfi, M.; Zarrabi, A.; Bigham, A.; Sharifi, E.; Rabiee, N.; Paiva-Santos, A.C.; Kumar, A.P.; Tan, S.C.; Hushmandi, K.; et al. Stimuli-responsive liposomal nanoformulations in cancer therapy: Pre-clinical & clinical approaches. J. Control. Release 2022, 351, 50–80. [Google Scholar] [PubMed]
- Nujoom, N.; Koyakutty, M.; Biswas, L.; Rajkumar, T.; Nair, S.V. Emerging Gene-editing nano-therapeutics for Cancer. Heliyon 2024, 10, e39323. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Gao, Y.; Zhang, L.; Zhang, Z.; Gao, F.; Ji, X. A pH-responsive mesoporous silica nanoparticles-based multi-drug delivery system for overcoming multi-drug resistance. Biomaterials 2011, 32, 7711–7720. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shahi, P.K.; Xie, R.; Zhang, H.; Abdeen, A.A.; Yodsanit, N. A pH-responsive silica–metal–organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, CRISPR-Cas9 genome-editing machineries. J. Control. Release 2020, 324, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Bazban-Shotorbani, S.; Hasani-Sadrabadi, M.M.; Karkhaneh, A.; Serpooshan, V.; Jacob, K.I.; Moshaverinia, A.; Mahmoudi, M. Revisiting structure-property relationship of pH-responsive polymers for drug delivery applications. J. Control. Release 2017, 253, 46–63. [Google Scholar] [CrossRef]
- Wang, C.-S.; Chang, C.-H.; Tzeng, T.-Y.; Lin, A.M.-Y.; Lo, Y.-L. Gene-editing by CRISPR–Cas9 in combination with anthracycline therapy via tumor microenvironment-switchable, EGFR-targeted, nucleus-directed nanoparticles for head and neck cancer suppression. Nanoscale Horiz. 2021, 6, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Lang, T.; Cun, D.; Zheng, Z.; Huang, Y.; Yin, Q. pH-sensitive nano-complexes overcome drug resistance and inhibit metastasis of breast cancer by silencing Akt expression. Theranostics 2017, 7, 4204. [Google Scholar] [CrossRef]
- Allemailem, K.S.; Almatroodi, S.A.; Almatroudi, A.; Alrumaihi, F.; Al Abdulmonem, W.; Al-Megrin, W.A.I.; Aljamaan, A.N.; Rahmani, A.H.; Khan, A.A. Recent advances in genome-editing technology with CRISPR/Cas9 variants and stimuli-responsive targeting approaches within tumor cells: A future perspective of cancer management. Int. J. Mol. Sci. 2023, 24, 7052. [Google Scholar] [CrossRef]
- Qian, Y.; Wang, X.; Li, Y.; Cao, Y.; Chen, X. Extracellular ATP a new player in cancer metabolism: NSCLC cells internalize ATP in vitro and in vivo using multiple endocytic mechanisms. Mol. Cancer Res. 2016, 14, 1087–1096. [Google Scholar] [CrossRef]
- Yang, X.; Tang, Q.; Jiang, Y.; Zhang, M.; Wang, M.; Mao, L. Nanoscale ATP-responsive zeolitic imidazole framework-90 as a general platform for cytosolic protein delivery and genome editing. J. Am. Chem. Soc. 2019, 141, 3782–3786. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, N.; Ishii, T.; Naito, M.; Endo, T.; Uchida, S.; Cabral, H.; Osada, K.; Kataoka, K. Polyplex micelles with phenylboronate/gluconamide cross-linking in the core exerting promoted gene transfection through spatiotemporal responsivity to intracellular pH and ATP concentration. J. Am. Chem. Soc. 2017, 139, 18567–18575. [Google Scholar] [CrossRef]
- Fan, X.; Zhao, X.; Su, W.; Tang, X. Triton X-100-Modified Adenosine Triphosphate-Responsive siRNA Delivery Agent for Antitumor Therapy. Mol. Pharm. 2020, 17, 3696–3708. [Google Scholar] [CrossRef] [PubMed]
- Giustarini, D.; Colombo, G.; Garavaglia, M.L.; Astori, E.; Portinaro, N.M.; Reggiani, F.; Badalamenti, S.; Aloisi, A.M.; Santucci, A.; Rossi, R.; et al. Assessment of glutathione/glutathione disulphide ratio and S-glutathionylated proteins in human blood, solid tissues, cultured cells. Free Radic. Biol. Med. 2017, 112, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Huo, M.; Yuan, J.; Tao, L.; Wei, Y. Redox-responsive polymers for drug delivery: From molecular design to applications. Polym. Chem. 2014, 5, 1519–1528. [Google Scholar] [CrossRef]
- Wang, Y.; Shahi, P.K.; Wang, X.; Xie, R.; Zhao, Y.; Wu, M. In vivo targeted delivery of nucleic acids and CRISPR genome editors enabled by GSH-responsive silica nanoparticles. J. Control. Release 2021, 336, 296–309. [Google Scholar] [CrossRef]
- Chen, G.; Abdeen, A.A.; Wang, Y.; Shahi, P.K.; Robertson, S.; Xie, R.; Suzuki, M.; Pattnaik, B.R.; Saha, K.; Gong, S. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol. 2019, 14, 974–980. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS sources in physiological and pathological conditions. Oxidative Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Pan, Q.; Xin, H.; Chen, Y.; Ping, Y. Genome-editing prodrug: Targeted delivery and conditional stabilization of CRISPR-Cas9 for precision therapy of inflammatory disease. Sci. Adv. 2021, 7, eabj0624. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, Y.; Chen, C.; Gao, Y.; Liu, X.; Yang, K.; Luan, X.; Zhou, D.; Zeng, F.; Han, X.; et al. Hypoxia-Responsive Gene Editing to Reduce Tumor Thermal Tolerance for Mild-Photothermal Therapy. Angew. Chem. Int. Ed. Engl. 2021, 60, 21200–21204. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Fan, B.; Sun, P.; Huang, W.; Jin, M.; Wang, Q.; Gao, Z. An effective tumor-targeting strategy utilizing hypoxia-sensitive siRNA delivery system for improved anti-tumor outcome. Acta Biomater. 2016, 44, 341–354. [Google Scholar] [CrossRef] [PubMed]
- De La Rica, R.; Aili, D.; Stevens, M.M. Enzyme-responsive nanoparticles for drug release and diagnostics. Adv. Drug Deliv. Rev. 2012, 64, 967–978. [Google Scholar] [CrossRef]
- Mu, J.; Lin, J.; Huang, P.; Chen, X. Development of endogenous enzyme-responsive nanomaterials for theranostics. Chem. Soc. Rev. 2018, 47, 5554–5573. [Google Scholar] [CrossRef]
- Simpson, M.A.; Lokeshwar, V.B. Hyaluronan and hyaluronidase in genitourinary tumors. Front. Biosci. A J. Virtual Libr. 2008, 13, 5664. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hou, S.; Wang, Q.; Bao, L.; Liu, D.; Yue, Y.; Yao, W.; Gao, X. Microenvironment-responsive delivery of the Cas9 RNA-guided endonuclease for efficient genome editing. Bioconjugate Chem. 2019, 30, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Song, L.; Liu, X.; Yang, X.; Li, X.; He, T.; Wang, N.; Yang, S.; Yu, C.; Yin, T.; et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 2017, 11, 95–111. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Guo, S.; Wu, L.; Chen, P.; Wang, L.; Liu, Y.; Ju, H. Near-infrared boosted ROS responsive siRNA delivery and cancer therapy with sequentially peeled upconversion nano-onions. Biomaterials 2019, 225, 119501. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.-M.; Tsang, F.H.-C.; Ng, I.-L.O. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, X.; Li, Y.; Wang, D.; Liu, W.; Zhang, Z.; Liu, J.; Zhang, K. MicroRNA-responsive release of Cas9/sgRNA from DNA nanoflower for cytosolic protein delivery and enhanced genome editing. Biomaterials 2020, 256, 120221. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Y.; Pan, W.; Yu, Z.; Yang, L.; Wang, H.; Li, N.; Tang, B. Nanocarriers with multi-locked DNA valves targeting intracellular tumor-related mRNAs for controlled drug release. Nanoscale 2017, 9, 17318–17324. [Google Scholar] [CrossRef] [PubMed]
- Bawa, P.; Pillay, V.; Choonara, Y.E.; Du Toit, L.C. Stimuli-responsive polymers and their applications in drug delivery. Biomed. Mater. 2009, 4, 22001. [Google Scholar] [CrossRef]
- Chen, C.; Ma, Y.; Du, S.; Wu, Y.; Shen, P.; Yan, T.; Li, X.; Song, Y.; Zha, Z.; Han, X. Controlled CRISPR-Cas9 ribonucleoprotein delivery for sensitized photothermal therapy. Small 2021, 17, 2101155. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, X.; Guan, N.; Shao, J.; Li, H.; Chen, Y.; Ping, Y.; Li, D.; Ye, H. Engineering a far-red light–activated split-Cas9 system for remote-controlled genome editing of internal organs and tumors. Sci. Adv. 2020, 6, eabb1777. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Cao, Y.; Tang, Y.; Yang, X.; Liu, Y.; Huang, D.; Zhang, Y.; Li, C.; Wang, Q. Advanced near-infrared light for monitoring and modulating the spatiotemporal dynamics of cell functions in living systems. Adv. Sci. 2020, 7, 1903783. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, Z.; Zhu, S.; He, L.; Fan, W.; Tang, W. A Rationally Designed Semiconducting Polymer Brush for NIR-II imaging-guided light-triggered remote control of CRISPR/Cas9 genome editing. Adv. Mater. 2019, 31, 1901187. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Zheng, W.; Cong, L.; Guo, Z.; Xie, Y.; Wang, L.; Tang, R.; Feng, Q.; Hamada, Y.; et al. Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem. Int. Ed. 2018, 57, 1491–1496. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, X.; Liu, S.; Chen, J.; Li, M.; Chew, S.Y.; Leong, K.W.; Cheng, D. Codelivery of CRISPR-Cas9 and chlorin e6 for spatially controlled tumor-specific gene editing with synergistic drug effects. Sci. Adv. 2020, 6, eabb4005. [Google Scholar] [CrossRef]
- Pan, Y.; Yang, J.; Luan, X.; Liu, X.; Li, X.; Yang, J.; Huang, T.; Sun, L.; Wang, Y.; Lin, Y.; et al. Near-infrared upconversion-activated CRISPR-Cas9 system: A remote-controlled gene editing platform. Sci. Adv. 2019, 5, eaav7199. [Google Scholar] [CrossRef]
- Perolina, E.; Meissner, S.; Raos, B.; Harland, B.; Thakur, S.; Svirskis, D. Translating ultrasound-mediated drug delivery technologies for CNS applications. Adv. Drug Deliv. Rev. 2024, 208, 115274. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Hynynen, K.; Lipsman, N. Applications of focused ultrasound in the brain: From thermoablation to drug delivery. Nat. Rev. Neurol. 2021, 17, 7–22. [Google Scholar] [CrossRef]
- Yang, Q.; Zhou, Y.; Chen, J.; Huang, N.; Wang, Z.; Cheng, Y. Gene therapy for drug-resistant glioblastoma via lipid-polymer hybrid nanoparticles combined with focused ultrasound. Int. J. Nanomed. 2021, 16, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.-Y.; Won, E.-J.; Lee, H.A.R.; Kim, J.H.; Hui, E.; Kim, H.P.; Yoon, T.-J. Ultrasound-activated particles as CRISPR/Cas9 delivery system for androgenic alopecia therapy. Biomaterials 2020, 232, 119736. [Google Scholar] [CrossRef]
- Kumar, C.S.; Mohammad, F. Magnetic nanomaterials for hyperthermia-based therapy and controlled drug delivery. Adv. Drug Deliv. Rev. 2011, 63, 789–808. [Google Scholar] [CrossRef]
- Thevenot, J.; Oliveira, H.; Sandre, O.; Lecommandoux, S. Magnetic responsive polymer composite materials. Chem. Soc. Rev. 2013, 42, 7099–7116. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Ghasemi, A.; Zangabad, P.S.; Rahighi, R.; Basri, S.M.M.; Mirshekari, H.; Amiri, M.; Pishabad, Z.S.; Aslani, A.; Bozorgomid, M.; et al. Smart micro/nanoparticles in stimulus-responsive drug/gene delivery systems. Chem. Soc. Rev. 2016, 45, 1457–1501. [Google Scholar] [CrossRef] [PubMed]
- Hryhorowicz, M.; Grześkowiak, B.; Mazurkiewicz, N.; Śledziński, P.; Lipiński, D.; Słomski, R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol. Biotechnol. 2019, 61, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.; Yndart, A.; Atluri, V.; Tiwari, S.; Tomitaka, A.; Gupta, P. Magnetically guided non-invasive CRISPR-Cas9/gRNA delivery across blood-brain barrier to eradicate latent HIV-1 infection. Sci. Rep. 2019, 9, 3928. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef]
- Nymark, M.; Sharma, A.K.; Sparstad, T.; Bones, A.M.; Winge, P. A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 2016, 6, 24951. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Yin, H.; Xue, W.; Chen, S.; Bogorad, R.L.; Benedetti, E.; Grompe, M.; Koteliansky, V.; A Sharp, P.; Jacks, T.; Anderson, D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014, 32, 551–553. [Google Scholar] [CrossRef]
- Demirci, S.; Leonard, A.; Haro-Mora, J.J.; Uchida, N.; Tisdale, J.F. CRISPR/Cas9 for sickle cell disease: Applications, future possibilities, challenges. Cell Biol. Transl. Med. Stem Cells: Transl. Sci. Ther. 2019, 5, 37–52. [Google Scholar]
- Brandow, A.; Liem, R. Advances in the diagnosis and treatment of sickle cell disease. J. Hematol. Oncol. 2022, 15, 20. [Google Scholar] [CrossRef] [PubMed]
- Aimo, A.; Castiglione, V.; Rapezzi, C.; Franzini, M.; Panichella, G.; Vergaro, G.; Gillmore, J.; Fontana, M.; Passino, C.; Emdin, M. RNA-targeting and gene editing therapies for transthyretin amyloidosis. Nat. Rev. Cardiol. 2022, 19, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Ruan, G.-X.; Barry, E.; Yu, D.; Lukason, M.; Cheng, S.H.; Scaria, A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber congenital amaurosis 10. Mol. Ther. 2017, 25, 331–341. [Google Scholar] [CrossRef]
- Lorenzo-Redondo, R.; Fryer, H.R.; Bedford, T.; Kim, E.Y.; Archer, J.; Kosakovsky Pond, S.L.K.; Chung, Y.S.; Penugonda, S.; Chipman, J.G.; Fletcher, C.V.; et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016, 530, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Young, C.S.; Hicks, M.R.; Ermolova, N.V.; Nakano, H.; Jan, M.; Younesi, S. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell 2016, 18, 533–540. [Google Scholar] [CrossRef]
- Wei, X.; Pu, A.; Liu, Q.; Hou, Q.; Zhang, Y.; An, X.; Long, Y.; Jiang, Y.; Dong, Z.; Wu, S.; et al. The bibliometric landscape of gene editing innovation and regulation in the worldwide. Cells 2022, 11, 2682. [Google Scholar] [CrossRef] [PubMed]
- Vangah, S.J.; Katalani, C.; Boone, H.A.; Hajizade, A.; Sijercic, A.; Ahmadian, G. CRISPR-based diagnosis of infectious and noninfectious diseases. Biol. Proced. Online 2020, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Uddin, F.; Rudin, T.; Sen, C.M. CRISPR gene therapy: Applications, limitations, implications for the future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef]
- He, S. The first human trial of CRISPR-based cell therapy clears safety concerns as new treatment for late-stage lung cancer. Signal Transduct. Target. Ther. 2020, 5, 168. [Google Scholar] [CrossRef]
- Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. Methodologies for improving HDR efficiency. Front. Genet. 2019, 9, 691. [Google Scholar] [CrossRef]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.J.; Sun, L.; Yao, Y.; Zhang, C.-Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 2021, 53, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021, 39, 35–40. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, B.; Chen, L.; Xie, L.; Yu, W.; Wang, Y.; Li, L.; Yin, S.; Yang, L.; Hu, H.; et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020, 38, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef]
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.F. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021, 184, 5635–5652. [Google Scholar] [CrossRef]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of plant prime-editing systems for precise genome editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef]
- Zhao, Z.; Shang, M.; Geijsen, N. Prime editing: Advances and therapeutic applications. Trends Biotechnol. 2023, 41, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.J.; Liu, D.R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2023, 24, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Newby, G.A.; Liu, D.R. In vivo somatic cell base editing and prime editing. Mol. Ther. 2021, 29, 3107–3124. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Rho, W.-Y.; Kim, Y.-H.; Chang, H.; Jun, B.-H. CRISPR-Cas9 Gene Therapy: Non-Viral Delivery and Stimuli-Responsive Nanoformulations. Molecules 2025, 30, 542. https://doi.org/10.3390/molecules30030542
Lee H, Rho W-Y, Kim Y-H, Chang H, Jun B-H. CRISPR-Cas9 Gene Therapy: Non-Viral Delivery and Stimuli-Responsive Nanoformulations. Molecules. 2025; 30(3):542. https://doi.org/10.3390/molecules30030542
Chicago/Turabian StyleLee, Hyunwoo, Won-Yeop Rho, Yoon-Hee Kim, Hyejin Chang, and Bong-Hyun Jun. 2025. "CRISPR-Cas9 Gene Therapy: Non-Viral Delivery and Stimuli-Responsive Nanoformulations" Molecules 30, no. 3: 542. https://doi.org/10.3390/molecules30030542
APA StyleLee, H., Rho, W.-Y., Kim, Y.-H., Chang, H., & Jun, B.-H. (2025). CRISPR-Cas9 Gene Therapy: Non-Viral Delivery and Stimuli-Responsive Nanoformulations. Molecules, 30(3), 542. https://doi.org/10.3390/molecules30030542







