pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Spectroscopic Analysis
2.3. Crystallization
2.4. Data Collection
2.5. Structure Determination
2.6. Bioinformatics
3. Results
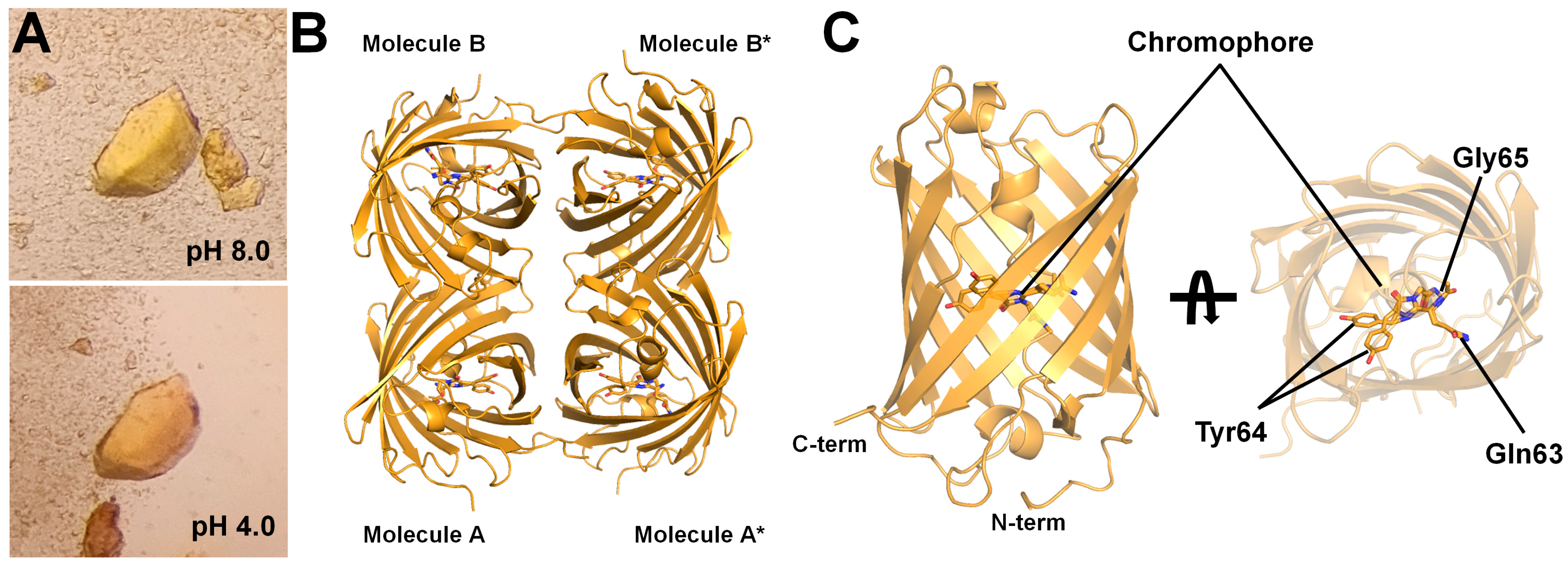
3.1. Spectroscopic Analysis of tKeima
3.2. Data Collection and Determination of tKeima Structure
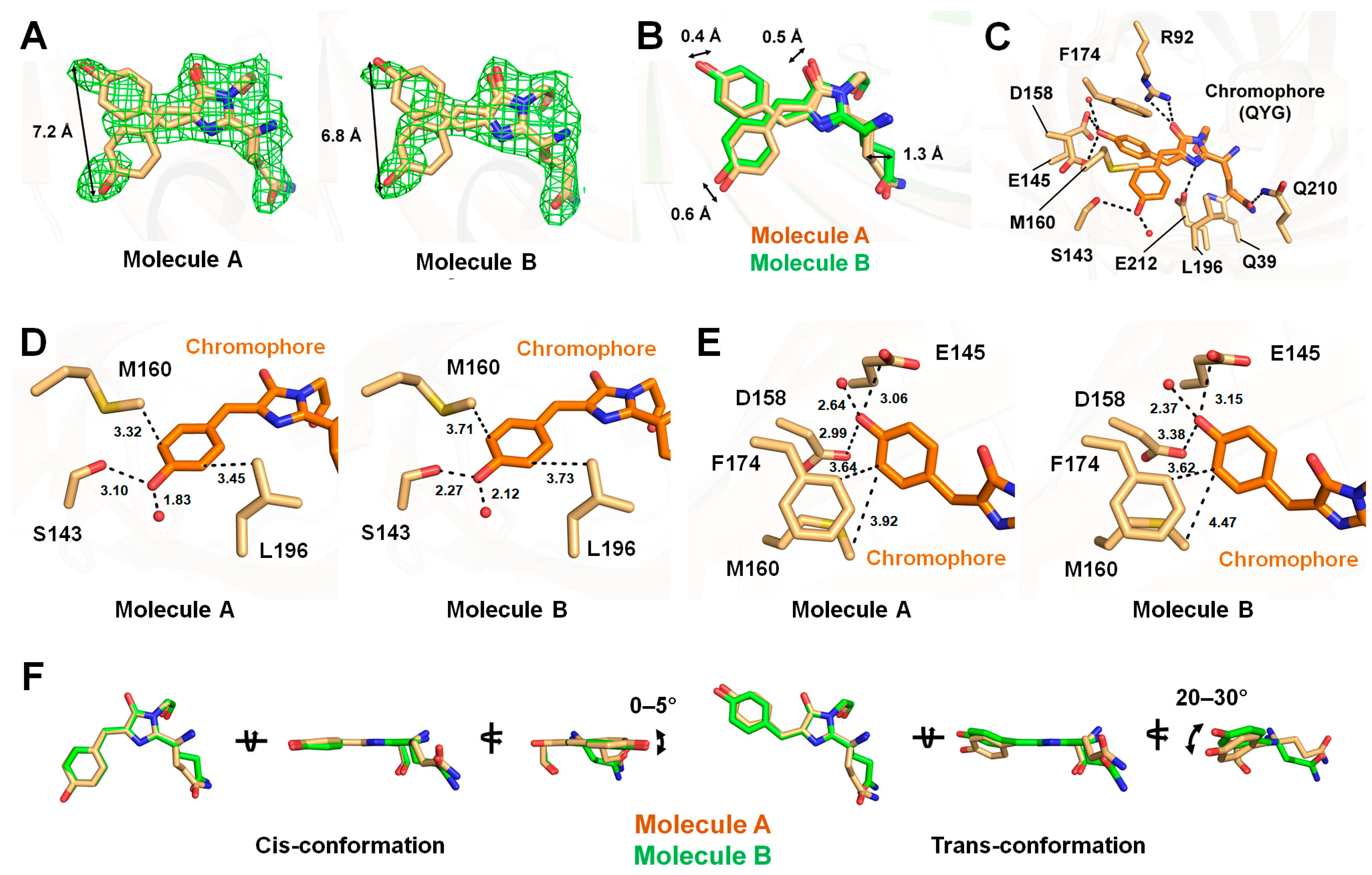
3.3. Chromophore of tKeimapH4.0
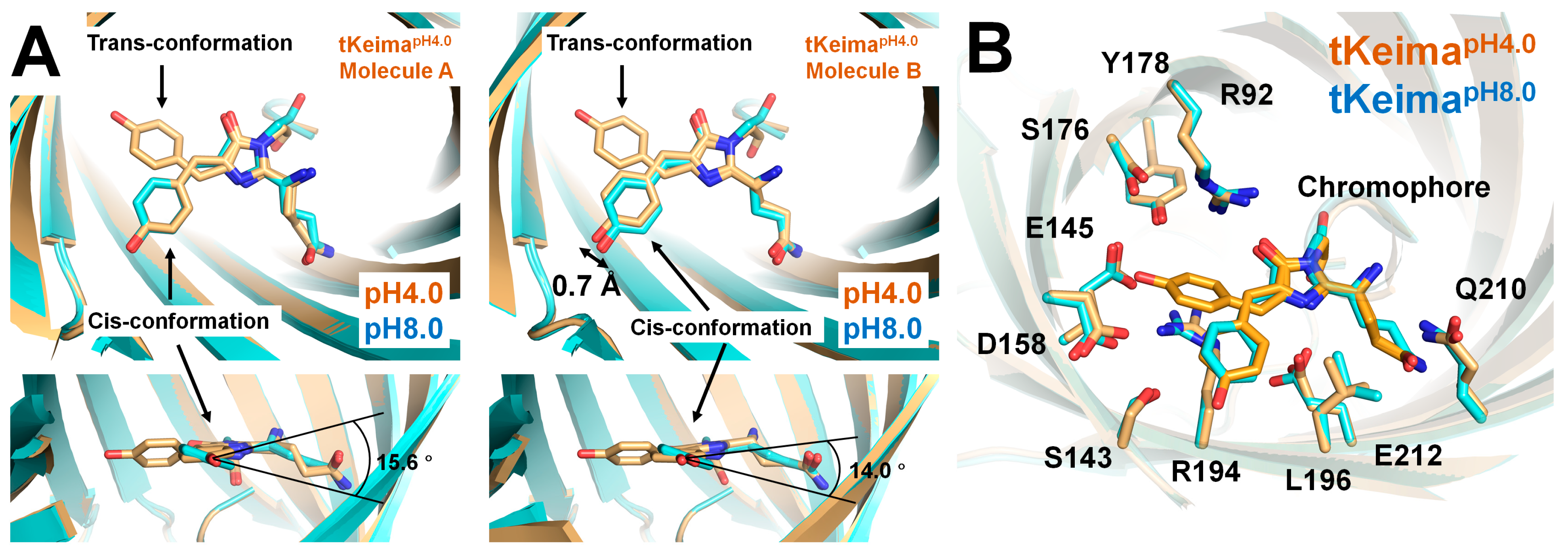
3.4. Comparison Between tKeimapH4.0 and tKeimapH8.0
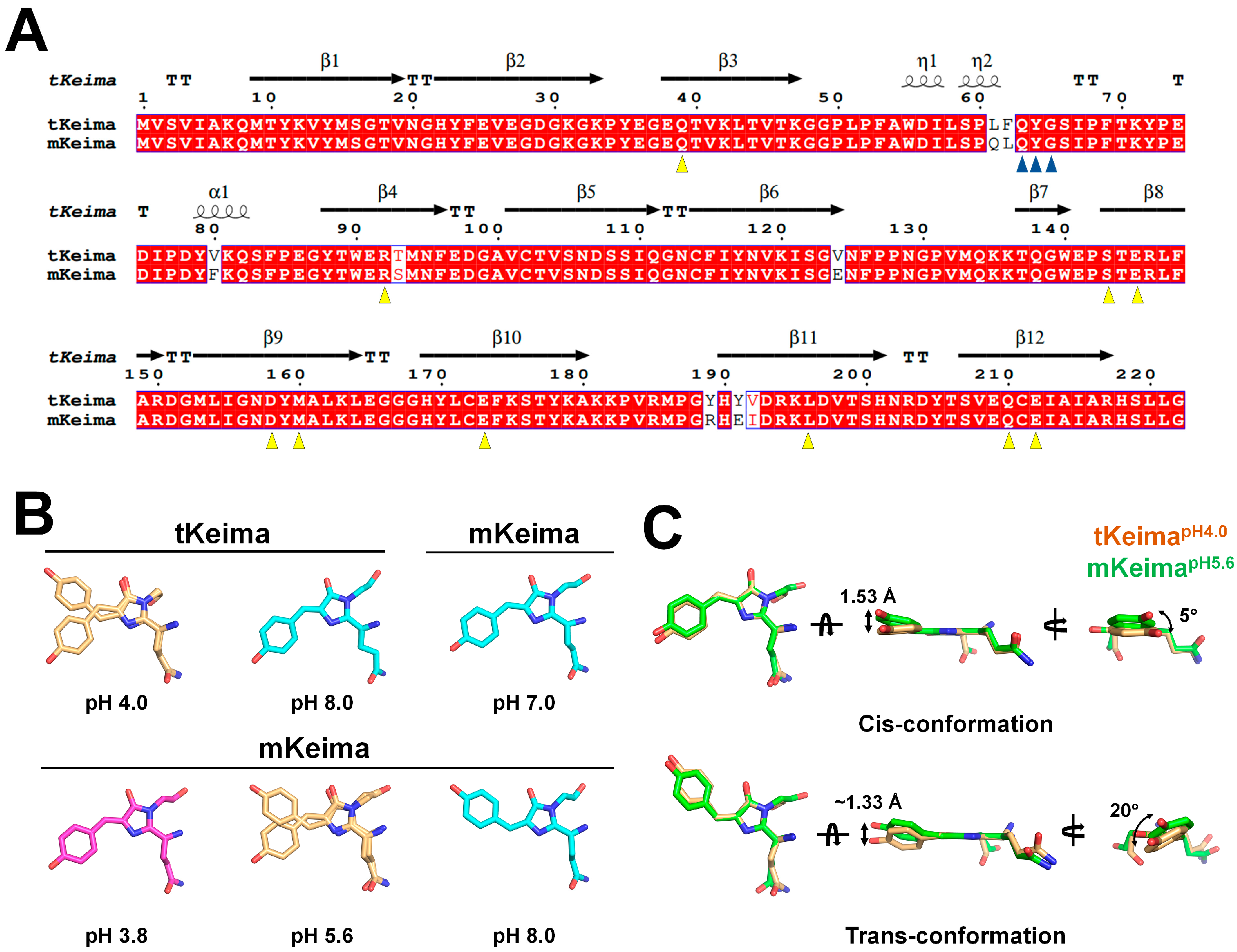
3.5. Comparison with Other Keima Families
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Sample, V.; Newman, R.H.; Zhang, J. The structure and function of fluorescent proteins. Chem. Soc. Rev. 2009, 38, 2852–2864. [Google Scholar] [CrossRef] [PubMed]
- Remington, S.J. Green fluorescent protein: A perspective. Protein Sci. 2011, 20, 1509–1519. [Google Scholar] [CrossRef]
- Tanz, S.K.; Castleden, I.; Small, I.D.; Millar, A.H. Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front. Plant Sci. 2013, 4, 214. [Google Scholar] [CrossRef]
- Cui, Y.; Gao, C.; Zhao, Q.; Jiang, L. Using Fluorescent Protein Fusions to Study Protein Subcellular Localization and Dynamics in Plant Cells. In High-Resolution Imaging of Cellular Proteins; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2016; pp. 113–123. [Google Scholar]
- Hynes, T.R.; Hughes, T.E.; Berlot, C.H. Cellular Localization of GFP-Tagged α Subunits. Methods Mol. Biol. 2004, 237, 233–246. [Google Scholar] [PubMed]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Liao, J.; Madahar, V.; Dang, R.; Jiang, L. Quantitative FRET (qFRET) Technology for the Determination of Protein–Protein Interaction Affinity in Solution. Molecules 2021, 26, 6339. [Google Scholar] [CrossRef]
- Skruzny, M.; Pohl, E.; Abella, M. FRET Microscopy in Yeast. Biosensors 2019, 9, 122. [Google Scholar] [CrossRef]
- Chudakov, D.M.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005, 23, 605–613. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J. Emerging In Vivo Analyses of Cell Function Using Fluorescence Imaging. Annu. Rev. Biochem. 2011, 80, 327–332. [Google Scholar] [CrossRef]
- Borg, R.E.; Rochford, J. Molecular Photoacoustic Contrast Agents: Design Principles & Applications. Photochem. Photobiol. 2018, 94, 1175–1209. [Google Scholar] [CrossRef] [PubMed]
- Galietta, L.J.V.; Haggie, P.M.; Verkman, A.S. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001, 499, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Bizzarri, R.; Arcangeli, C.; Arosio, D.; Ricci, F.; Faraci, P.; Cardarelli, F.; Beltram, F. Development of a Novel GFP-based Ratiometric Excitation and Emission pH Indicator for Intracellular Studies. Biophys. J. 2006, 90, 3300–3314. [Google Scholar] [CrossRef] [PubMed]
- Frommer, W.B.; Davidson, M.W.; Campbell, R.E. Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 2009, 38, 2833. [Google Scholar] [CrossRef]
- Nam, K.H. Fluorescent Protein-Based Metal Biosensors. Chemosensors 2023, 11, 216. [Google Scholar] [CrossRef]
- Perez-Rodriguez, R.; Haitjema, C.; Huang, Q.; Nam, K.H.; Bernardis, S.; Ke, A.; DeLisa, M.P. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 2011, 79, 584–599. [Google Scholar] [CrossRef]
- Dolan, A.E.; Hou, Z.; Xiao, Y.; Gramelspacher, M.J.; Heo, J.; Howden, S.E.; Freddolino, P.L.; Ke, A.; Zhang, Y. Introducing a Spectrum of Long-Range Genomic Deletions in Human Embryonic Stem Cells Using Type I CRISPR-Cas. Mol. Cell. 2019, 74, 936–950.e5. [Google Scholar] [CrossRef]
- Hu, C.; Ni, D.; Nam, K.H.; Majumdar, S.; McLean, J.; Stahlberg, H.; Terns, M.P.; Ke, A. Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools. Mol. Cell. 2022, 82, 2754–2768.e5. [Google Scholar] [CrossRef]
- Tan, R.; Krueger, R.K.; Gramelspacher, M.J.; Zhou, X.; Xiao, Y.; Ke, A.; Hou, Z.; Zhang, Y. Cas11 enables genome engineering in human cells with compact CRISPR-Cas3 systems. Mol. Cell. 2022, 82, 852–867.e5. [Google Scholar] [CrossRef]
- Nienhaus, G.U.; Wiedenmann, J. Structure, Dynamics and Optical Properties of Fluorescent Proteins: Perspectives for Marker Development. ChemPhysChem 2009, 10, 1369–1379. [Google Scholar] [CrossRef]
- Kim, I.J.; Kim, S.; Park, J.; Eom, I.; Kim, S.; Kim, J.H.; Ha, S.C.; Kim, Y.G.; Hwang, K.Y.; Nam, K.H. Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development. FEBS Lett. 2016, 590, 2982–2990. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Metal-Induced Fluorescence Quenching of Photoconvertible Fluorescent Protein DendFP. Molecules 2022, 27, 2922. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.V.; Solá, H.M.; Torres, J.C.; Torres, R.E.; Guzmán, E.E. Effect of pH on the Heat-Induced Denaturation and Renaturation of Green Fluorescent Protein: A Laboratory Experiment. J. Chem. Educ. 2012, 90, 248–251. [Google Scholar] [CrossRef]
- Penna, T.C.V.; Ishii, M.; Junior, A.P.; Cholewa, O. Thermal Stability of Recombinant Green Fluorescent Protein (GFPuv) at Various pH Values. Appl. Biochem. Biotechnol. 2004, 114, 469–484. [Google Scholar] [CrossRef]
- Roberts, T.M.; Rudolf, F.; Meyer, A.; Pellaux, R.; Whitehead, E.; Panke, S.; Held, M. Identification and Characterisation of a pH-stable GFP. Sci. Rep. 2016, 6, 28166. [Google Scholar] [CrossRef]
- Liu, A.; Huang, X.; He, W.; Xue, F.; Yang, Y.; Liu, J.; Chen, L.; Yuan, L.; Xu, P. pHmScarlet is a pH-sensitive red fluorescent protein to monitor exocytosis docking and fusion steps. Nat. Commun. 2021, 12, 1413. [Google Scholar] [CrossRef]
- Kneen, M.; Farinas, J.; Li, Y.; Verkman, A.S. Green Fluorescent Protein as a Noninvasive Intracellular pH Indicator. Biophys. J. 1998, 74, 1591–1599. [Google Scholar] [CrossRef]
- Han, J.; Burgess, K. Fluorescent Indicators for Intracellular pH. Chem. Rev. 2009, 110, 2709–2728. [Google Scholar] [CrossRef]
- Kim, H.; Lee, W.; Yoon, Y. Heavy metal(loid) biosensor based on split-enhanced green fluorescent protein: Development and characterization. Appl. Microbiol. Biotechnol. 2019, 103, 6345–6352. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Spectroscopic and Structural Analysis of Cu(2+)-Induced Fluorescence Quenching of ZsYellow. Biosensors 2020, 10, 29. [Google Scholar] [CrossRef]
- Oltrogge, L.M.; Wang, Q.; Boxer, S.G. Ground-State Proton Transfer Kinetics in Green Fluorescent Protein. Biochemistry 2014, 53, 5947–5957. [Google Scholar] [CrossRef] [PubMed]
- Scharnagl, C.; Raupp-Kossmann, R.; Fischer, S.F. Molecular Basis for pH Sensitivity and Proton Transfer in Green Fluorescent Protein: Protonation and Conformational Substates from Electrostatic Calculations. Biophys. J. 1999, 77, 1839–1857. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Byrnes, L.J.; Shui, B.; Röhrig, U.F.; Singh, A.; Chudakov, D.M.; Lukyanov, S.; Zipfel, W.R.; Kotlikoff, M.I.; Sondermann, H. Molecular Mechanism of a Green-Shifted, pH-Dependent Red Fluorescent Protein mKate Variant. PLoS ONE 2011, 6, e23513. [Google Scholar] [CrossRef]
- Bae, J.E.; Kim, I.J.; Nam, K.H. Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221. Biochem. Biophys. Res. Commun. 2017, 493, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.M.; Sheng, W.; Esmatpour Salmani, R.; Tahmasebi Nick, S.; Ghanbarpour, A.; Gholami, H.; Vasileiou, C.; Geiger, J.H.; Borhan, B. Design of Large Stokes Shift Fluorescent Proteins Based on Excited State Proton Transfer of an Engineered Photobase. J. Am. Chem. Soc. 2021, 143, 15091–15102. [Google Scholar] [CrossRef]
- Kogure, T.; Karasawa, S.; Araki, T.; Saito, K.; Kinjo, M.; Miyawaki, A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006, 24, 577–581. [Google Scholar] [CrossRef]
- Kogure, T.; Kawano, H.; Abe, Y.; Miyawaki, A. Fluorescence imaging using a fluorescent protein with a large Stokes shift. Methods 2008, 45, 223–226. [Google Scholar] [CrossRef]
- Day, R.N.; Davidson, M.W. The fluorescent protein palette: Tools for cellular imaging. Chem. Soc. Rev. 2009, 38, 2887. [Google Scholar] [CrossRef]
- Ma, X.; Foo, Y.H.; Wohland, T. Fluorescence Cross-Correlation Spectroscopy (FCCS) in Living Cells. In Fluorescence Spectroscopy and Microscopy; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2014; pp. 557–573. [Google Scholar]
- Sun, N.; Malide, D.; Liu, J.; Rovira, I.I.; Combs, C.A.; Finkel, T. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat. Protoc. 2017, 12, 1576–1587. [Google Scholar] [CrossRef]
- Gilsrud, A.J.; Narendra, D.P. Methods for Monitoring Mitophagy Using mt-Keima. In Selective Autophagy; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2024; pp. 151–160. [Google Scholar]
- Karantanou, C.; Bibli, S.-I. Monitoring Mitophagy Dynamics in Live Cells. In Selective Autophagy; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2024; pp. 161–175. [Google Scholar]
- Im, S.; Jeong, D.J.; Kim, E.; Choi, J.H.; Jang, H.J.; Kim, Y.Y.; Um, J.H.; Lee, J.; Lee, Y.J.; Lee, K.M.; et al. A novel marine-derived mitophagy inducer ameliorates mitochondrial dysfunction and thermal hypersensitivity in paclitaxel-induced peripheral neuropathy. Br. J. Pharmacol. 2024, 181, 4012–4027. [Google Scholar] [CrossRef]
- Yang, Y.; Ke, J.; Cao, Y.; Gao, Y.; Lin, C. Melatonin regulates microglial M1/M2 polarization via AMPKα2-mediated mitophagy in attenuating sepsis-associated encephalopathy. Biomed. Pharmacother. 2024, 177, 117092. [Google Scholar] [CrossRef]
- Nam, K.H.; Xu, Y. Structural Analysis of the Large Stokes Shift Red Fluorescent Protein tKeima. Molecules 2024, 29, 2579. [Google Scholar] [CrossRef]
- Gu, D.H.; Eo, C.; Hwangbo, S.A.; Ha, S.C.; Kim, J.H.; Kim, H.; Lee, C.S.; Seo, I.D.; Yun, Y.D.; Lee, W.; et al. BL-11C Micro-MX: A high-flux microfocus macromolecular-crystallography beamline for micrometre-sized protein crystals at Pohang Light Source II. J. Synchrotron Radiat. 2021, 28, 1210–1215. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Metoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]
- Violot, S.; Carpentier, P.; Blanchoin, L.; Bourgeois, D. Reverse pH-Dependence of Chromophore Protonation Explains the Large Stokes Shift of the Red Fluorescent Protein mKeima. J. Am. Chem. Soc. 2009, 131, 10356–10357. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.N.; Osborn, M.F.; Koon, N.; Gepshtein, R.; Huppert, D.; Remington, S.J. Excited State Proton Transfer in the Red Fluorescent Protein mKeima. J. Am. Chem. Soc. 2009, 131, 13212–13213. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Structural Flexibility of the Monomeric Red Fluorescent Protein DsRed. Crystals 2024, 14, 62. [Google Scholar] [CrossRef]
- Adam, V.; Lelimousin, M.; Boehme, S.; Desfonds, G.; Nienhaus, K.; Field, M.J.; Wiedenmann, J.; McSweeney, S.; Nienhaus, G.U.; Bourgeois, D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc. Natl. Acad. Sci. USA 2008, 105, 18343–18348. [Google Scholar] [CrossRef]
- Henderson, J.N.; Ai, H.-w.; Campbell, R.E.; Remington, S.J. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc. Natl. Acad. Sci. USA 2007, 104, 6672–6677. [Google Scholar] [CrossRef]
- Luin, S.; Voliani, V.; Lanza, G.; Bizzarri, R.; Amat, P.; Tozzini, V.; Serresi, M.; Beltram, F. Raman Study of Chromophore States in Photochromic Fluorescent Proteins. J. Am. Chem. Soc. 2008, 131, 96–103. [Google Scholar] [CrossRef]

| Data Collection | tKeimapH4.0 |
|---|---|
| Synchrotron | Beamline 11, PLS-II |
| Wavelength (Å) | 0.9794 |
| Space group | P21212 |
| Unit cell | |
| a, b, c (Å) | 70.005, 85.773, 109.759 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0–2.20 (2.24–2.20) |
| Unique reflections | 34,083 (1658) |
| Completeness (%) | 99.0 (98.9) |
| Redundancy | 7.9 (6.3) |
| Mean I/σ(I) | 14.0 (2.58) |
| Rmerge | 0.131 (0.486) |
| CC1/2 | 0.998 (0.844) |
| CC* | 0.997 (0.957) |
| Refinement | |
| Resolution range (Å) | 46.23–2.20 (2.26–2.20) |
| Rwork | 0.2312 (0.3493) |
| Rfree | 0.2747 (0.3698) |
| R.m.s. deviations | |
| Bonds (Å) | 0.007 |
| Angles (°) | 1.104 |
| Average B factors (Å2) | |
| Protein | 27.42 |
| Water | 30.89 |
| Ramachandran plot (%) | |
| Most favored | 98.85 |
| Allowed | 1.15 |
| Outlier | 0 |
| PDB code | 9LPU |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Seo, Y.G.; Kim, I.J.; Nam, K.H. pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima. Molecules 2025, 30, 1623. https://doi.org/10.3390/molecules30071623
Xu Y, Seo YG, Kim IJ, Nam KH. pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima. Molecules. 2025; 30(7):1623. https://doi.org/10.3390/molecules30071623
Chicago/Turabian StyleXu, Yongbin, Yun Gyo Seo, In Jung Kim, and Ki Hyun Nam. 2025. "pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima" Molecules 30, no. 7: 1623. https://doi.org/10.3390/molecules30071623
APA StyleXu, Y., Seo, Y. G., Kim, I. J., & Nam, K. H. (2025). pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima. Molecules, 30(7), 1623. https://doi.org/10.3390/molecules30071623










