A Model of Interaction between Nicotinamide Adenine Dinucleotide Phosphate (NADPH) Oxidase and Apocynin Analogues by Docking Method
Abstract
:1. Introduction
2. Computational Methods
2.1. Database and Software
2.2. Preparation of Inhibitors
2.3. Identification of the Potential Active Site
2.4. Construction and Validation of the Docking Condition
3. Results and Discussion
3.1. Training Set and Test Set
3.2. Identification of the Potential Active Site
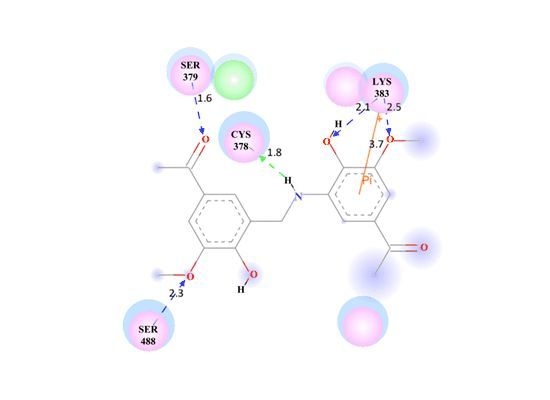
3.3. Molecular Docking of the Training Sets
3.4. Validation of the Hypothesis
4. Conclusions
Acknowledgements
References
- De Almeida, A.C.; Cabral, M.O.; Arslanian, C.; Condino-Neto, A.; Ximenes, V.F. 4-fluoro-2-methoxyphenol, an apocynin analog with enhanced inhibitory effect on leukocyte oxidant production and phagocytosis. Eur. J. Pharmacol 2011, 660, 445–453. [Google Scholar]
- Kanegae, M.P.; da Fonseca, L.M.; Brunetti, I.L.; Silva, S.O.; Ximenes, V.F. The reactivity of ortho-methoxy-substituted catechol radicals with sulfhydryl groups: Contribution for the comprehension of the mechanism of inhibition of NADPH oxidase by apocynin. Biochem. Pharmacol 2007, 74, 457–464. [Google Scholar]
- Johnson, J.L.; Park, J.W.; Benna, J.E.; Faust, L.P.; Inanami, O.; Babior, B.M. Activation of p47(PHOX), a cytosolic subunit of the leukocyte NADPH oxidase. Phosphorylation of ser-359 or ser-370 precedes phosphorylation at other sites and is required for activity. J. Biol. Chem 1998, 273, 35147–35152. [Google Scholar]
- Steffen, Y.; Vuillaume, G.; Stolle, K.; Roewer, K.; Lietz, M.; Schueller, J.; Lebrun, S.; Wallerath, T. Cigarette smoke and LDL cooperate in reducing nitric oxide bioavailability in endothelial cells via effects on both eNOS and NADPH oxidase. Nitric Oxide 2012, 27, 176–184. [Google Scholar]
- Cai, H.; Griendling, K.K.; Harrison, D.G. The vascular NAD(P)H oxidases as therapeutic targets in cardiovascular diseases. Trends Pharmacol. Sci 2003, 24, 471–478. [Google Scholar]
- Griendling, K.K.; Minieri, C.A.; Ollerenshaw, J.D.; Alexander, R.W. Angiotensin II stimulates NADH and NADPH oxidase activity in cultured vascular smooth muscle cells. Circ. Res 1994, 74, 1141–1148. [Google Scholar]
- Impellizzeri, D.; Esposito, E.; Mazzon, E.; Paterniti, I.; di Paola, R.; Bramanti, P.; Cuzzocrea, S. Effect of apocynin, a NADPH oxidase inhibitor, on acute lung inflammation. Biochem. Pharmacol 2011, 81, 636–648. [Google Scholar]
- Selemidis, S.; Sobey, C.G.; Wingler, K.; Schmidt, H.H.; Drummond, G.R. NADPH oxidases in the vasculature: Molecular features, roles in disease and pharmacological inhibition. Pharmacol. Ther 2008, 120, 254–291. [Google Scholar]
- Tossi, V.; Cassia, R.; Lamattina, L. Apocynin-induced nitric oxide production confers antioxidant protection in maize leaves. J. Plant Physiol 2009, 166, 1336–1341. [Google Scholar]
- Engels, F.; Renirie, B.F.; Hart, B.A.; Labadie, R.P.; Nijkamp, F.P. Effects of apocynin, a drug isolated from the roots of Picrorhiza kurroa, on arachidonic acid metabolism. FEBS Lett 1992, 305, 254–256. [Google Scholar]
- Valdecir, F.X.; Iguatemy, L.B.; Luiz, M.D.F. Apocynin and related methoxy-catechols as inhibitors of neutrophil-NADPH oxidase in LPS-activated whole blood. Res. J. Biol. Sci 2007, 2, 360–364. [Google Scholar]
- Lu, X.; Wan, S.; Jiang, J.; Jiang, X.; Yang, W.; Yu, P.; Xu, L.; Zhang, Z.; Zhang, G.; Shan, L.; et al. Synthesis and biological evaluations of novel apocynin analogues. Eur. J. Med. Chem 2011, 46, 2691–2698. [Google Scholar]
- Steffen, Y.; Gruber, C.; Schewe, T.; Sies, H. Mono-O-methylated flavanols and other flavonoids as inhibitors of endothelial NADPH oxidase. Arch. Biochem. Biophys 2008, 469, 209–219. [Google Scholar]
- Mora-Pale, M.; Weiwer, M.; Yu, J.; Linhardt, R.J.; Dordick, J.S. Inhibition of human vascular NADPH oxidase by apocynin derived oligophenols. Bioorg. Med. Chem 2009, 17, 5146–5152. [Google Scholar]
- Yu, J.; Weiwer, M.; Linhardt, R.J.; Dordick, J.S. The role of the methoxyphenol apocynin, a vascular NADPH oxidase inhibitor, as a chemopreventative agent in the potential treatment of cardiovascular diseases. Curr. Vasc. Pharmacol 2008, 6, 204–217. [Google Scholar]
- Ximenes, V.F.; Kanegae, M.P.; Rissato, S.R.; Galhiane, M.S. The oxidation of apocynin catalyzed by myeloperoxidase: Proposal for NADPH oxidase inhibition. Arch. Biochem. Biophys 2007, 457, 134–141. [Google Scholar]
- Kawahara, T.; Lambeth, J.D. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evol. Biol 2007, 7, 178. [Google Scholar]
- Inanami, O.; Johnson, J.L.; Babior, B.M. The leukocyte NADPH oxidase subunit p47PHOX: The role of the cysteine residues. Arch. Biochem. Biophys 1998, 350, 36–40. [Google Scholar]
- Xu, J.; Lu, Y.J.; Shen, W.Z.; Luo, H.B.; Huang, S.C.; Bao, J.L.; Cai, S.H.; Wang, Y.Q. Homology modeling of α-glucosidase and its interactions with andrograpolide derivatives. Lett. Drug Des. Discov 2011, 8, 440–451. [Google Scholar]
- Doman, T.N.; McGovern, S.L.; Witherbee, B.J.; Kasten, T.P.; Kurumbail, R.; Stallings, W.C.; Connolly, D.T.; Shoichet, B.K. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J. Med. Chem 2002, 45, 2213–2221. [Google Scholar]
- Gehlhaar, D.K.; Verkhivker, G.M.; Rejto, P.A.; Sherman, C.J.; Fogel, D.B.; Fogel, L.J.; Freer, S.T. Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: Conformationally flexible docking by evolutionary programming. Chem. Biol 1995, 2, 317–324. [Google Scholar]
- DesJarlais, R.L.; Seibel, G.L.; Kuntz, I.D.; Furth, P.S.; Alvarez, J.C.; Ortiz de Montellano, P.R.; DeCamp, D.L.; Babe, L.M.; Craik, C.S. Structure-based design of nonpeptide inhibitors specific for the human immunodeficiency virus 1 protease. Proc. Natl. Acad. Sci. USA 1990, 87, 6644–6648. [Google Scholar]
- RCSB Protein Data BANK. Available online: http://www.rcsb.org/pdb/explore/explore.do?structureId=1K4U accessed on 5 September 2011.
- Verdonk, M.L.; Chessari, G.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Nissink, J.W.; Taylor, R.D.; Taylor, R. Modeling water molecules in protein-ligand docking using GOLD. J. Med. Chem 2005, 48, 6504–6515. [Google Scholar]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar]
- Halgren, T.A. Maximally diagonal force constants in dependent angle-bending coordinates. II. Implications for the design of empirical force fields. J. Am. Chem. Soc 1990, 112, 4710–4723. [Google Scholar]
- Clark, M.; Cramer, R.D., III; Opdenbosch, N.V. Validation of the general purpose tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar]
- Powell, M.J.D. Restart procedures for the conjugate gradient method. Math. Program 1977, 12, 241–254. [Google Scholar]
- Mannhold, R.; Rekker, R.F.; Sonntag, C.; ter Laak, A.M.; Dross, K.; Polymeropoulos, E.E. Comparative evaluation of the predictive power of calculation procedures for molecular lipophilicity. J. Pharm. Sci 1995, 84, 1410–1419. [Google Scholar]
- Ogura, K.; Nobuhisa, I.; Yuzawa, S.; Takeya, R.; Torikai, S.; Saikawa, K.; Sumimoto, H.; Inagaki, F. NMR solution structure of the tandem Src homology 3 domains of p47phox complexed with a p22phox-derived proline-rich peptide. J. Biol. Chem 2006, 281, 3660–3668. [Google Scholar]
- Groemping, Y.; Lapouge, K.; Smerdon, S.J.; Rittinger, K. Molecular basis of phosphorylation-induced activation of the NADPH oxidase. Cell 2003, 113, 343–355. [Google Scholar]
- Bissantz, C.; Folkers, G.; Rognan, D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J. Med. Chem 2000, 43, 4759–4767. [Google Scholar]





| Residues | |
|---|---|
| 1 | GLN461, VAL462, VAL486, LYS489, GLU492, TRP494, LEU495, LYS508, ASP531, SER460, THR516 |
| 2 | GLN461, VAL462, GLU463, ILE483, ILE484, LEU485, LYS458, SER514, SER460, ALA515 |
| 3 | PRO363, ALA364, PRO366, PRO367, ARG368, PRO369, ASN491, GLU492, GLU493, TRP494, LYS508 |
| 4 | GLN362, PRO363, ALA364, VAL365, PRO366, ARG368, SER467, TYR468, GLU469, GLN472, ASP475, TRP494, PHE510 |
| 5 | LYS385, ALA470, GLN472, PRO473, GLU474, ASP475, LEU476, GLU477, PHE478, GLN479, ASP482, ILE484, GLY497, SER499, LYS500, LYS502, VAL503, GLY504 |
| 6 | ARG377, CYS378, SER379, LEU487, SER488, LYS489, GLU496 |
| 7 | ARG368, PRO369, ALA371, ILE374, LYS385, LEU386, SER388, GLU474, ILE505 |
| Compound | NADPH oxidation | GOLD Score | Pi interaction | ||
|---|---|---|---|---|---|
| IC50a(μm)(Experimental) | |||||
| Training set | Predicted good | Apocynin dimer | 10.3 ± 2.5 | 45.0082 | - |
| Homovanillin alcohol | 5.1 ± 1.7 | 35.1074 | LYS383 | ||
| Tyrosol | 7.5 ± 2.4 | 32.8184 | LYS383 | ||
| Ferulic acid | 4.9 ± 1.6 | 31.7482 | LYS383 | ||
| Hydroxytyrosol | 7.5 ± 2.9 | 28.9217 | LYS383 | ||
| Caffeic acid | 15.0 ± 4.1 | 27.8341 | - | ||
| Predicted weak | 3-Methoxysalicylic acid | >100 | 27.3678 | - | |
| Apocynin | 50.0 ± 9.1 | 27.0364 | - | ||
| Vanillic acid | 8.1 ± 3.1 | 26.6326 | - | ||
| Homovanillin | 7.5 ± 2.6 | 26.0688 | SER379 | ||
| Acetylsalicylic acid | >100 | 25.7012 | - | ||
| Salicylic acid | >100 | 23.2333 | - | ||
| Test set | 12 | - | 52.0590 | LYS383 | |
| 6b | - | 42.8667 | - | ||
| 10 | - | 41.1901 | LYS383 | ||
| 6c | - | 40.7741 | - | ||
| 8c | - | 40.6892 | - | ||
| 3b | - | 38.6765 | - | ||
| 8a | - | 38.2455 | - | ||
| 8b | - | 37.7346 | - | ||
| 11 | - | 37.4821 | - | ||
| 6a | - | 36.7572 | - | ||
| 3a | - | 36.4118 | - | ||
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jiang, J.; Kang, H.; Song, X.; Huang, S.; Li, S.; Xu, J. A Model of Interaction between Nicotinamide Adenine Dinucleotide Phosphate (NADPH) Oxidase and Apocynin Analogues by Docking Method. Int. J. Mol. Sci. 2013, 14, 807-817. https://doi.org/10.3390/ijms14010807
Jiang J, Kang H, Song X, Huang S, Li S, Xu J. A Model of Interaction between Nicotinamide Adenine Dinucleotide Phosphate (NADPH) Oxidase and Apocynin Analogues by Docking Method. International Journal of Molecular Sciences. 2013; 14(1):807-817. https://doi.org/10.3390/ijms14010807
Chicago/Turabian StyleJiang, Jie, Hongjun Kang, Xiaoliang Song, Sichao Huang, Sha Li, and Jun Xu. 2013. "A Model of Interaction between Nicotinamide Adenine Dinucleotide Phosphate (NADPH) Oxidase and Apocynin Analogues by Docking Method" International Journal of Molecular Sciences 14, no. 1: 807-817. https://doi.org/10.3390/ijms14010807
APA StyleJiang, J., Kang, H., Song, X., Huang, S., Li, S., & Xu, J. (2013). A Model of Interaction between Nicotinamide Adenine Dinucleotide Phosphate (NADPH) Oxidase and Apocynin Analogues by Docking Method. International Journal of Molecular Sciences, 14(1), 807-817. https://doi.org/10.3390/ijms14010807