Abstract
RNA-binding proteins (RBPs) are pivotal regulators of all the steps of gene expression. RBPs govern gene regulation at the post-transcriptional level by virtue of their capacity to assemble ribonucleoprotein complexes on certain RNA structural elements, both in normal cells and in response to various environmental stresses. A rapid cellular response to stress conditions is triggered at the step of translation initiation. Two basic mechanisms govern translation initiation in eukaryotic mRNAs, the cap-dependent initiation mechanism that operates in most mRNAs, and the internal ribosome entry site (IRES)-dependent mechanism activated under conditions that compromise the general translation pathway. IRES elements are cis-acting RNA sequences that recruit the translation machinery using a cap-independent mechanism often assisted by a subset of translation initiation factors and various RBPs. IRES-dependent initiation appears to use different strategies to recruit the translation machinery depending on the RNA organization of the region and the network of RBPs interacting with the element. In this review we discuss recent advances in understanding the implications of RBPs on IRES-dependent translation initiation.
1. Introduction
RNA plays a central role in gene expression. Within the cell RNA molecules are associated to RNA-binding proteins (RBPs) forming dynamic ribonucleoprotein particles (RNPs) that affect all steps of RNA metabolism []. RBPs assemble on nascent and processed mRNAs, governing gene regulation at post-transcriptional level in health and disease. Indeed, mutations affecting the function of RBPs cause several diseases []. RBPs are major components of the cell that control transcription, splicing, catalytic processing, transport, localization, translation or RNA stability processes (Figure 1). These steps in the RNA lifespan are closely connected to each other, such that altering one of them will affect the others.
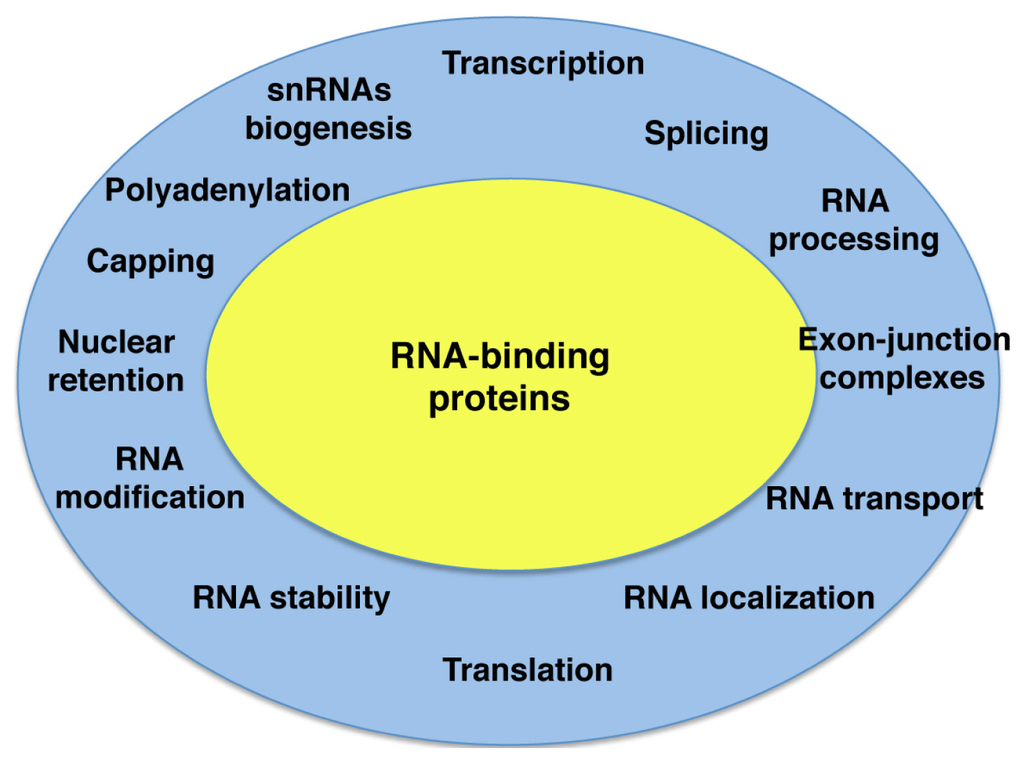
Figure 1.
Gene expression control in eukaryotic cells. Each step of gene expression is controlled by RNA-binding proteins (yellow), such that altering one step affects the rest (blue ring) impacting on translation efficiency of mRNAs.
RBPs often interact with the untranslated regions (UTRs) of mRNAs, which perform cis-acting regulatory functions in most cases and provide the landing site of many RBPs []. RBPs interacting with certain UTR structural elements or specific primary sequences play a pivotal role in the response of the cell to different environmental stresses, such as virus infections, heat or osmotic stress, nutrient deprivation and other stimuli that trigger apoptosis, inflammatory response, antiviral response, etc. (Figure 2) [–]. On the other hand, processes such as cell proliferation, cell death or cell differentiation occurring in healthy organisms also depend on RNA-protein interactions [].
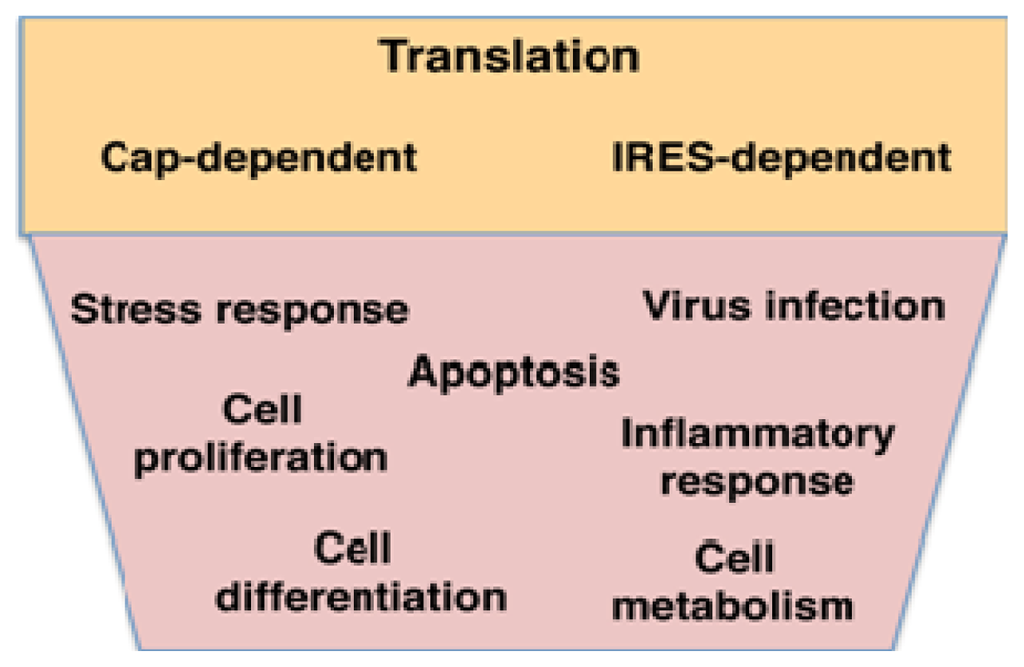
Figure 2.
RNA-binding proteins control translation efficiency of mRNAs initiating protein synthesis by the conventional cap-dependent, or the alternative internal ribosome entry site (IRES)-dependent, mechanisms (orange box), impacting on cellular processes in cells undergoing normal growth as well as in response to environmental stresses (pink box).
In response to distinct stresses, cells trigger a differential response that can displace the equilibrium towards cell survival or cell death. Key factors mediating this response are post-translational modification, relocalization, proteolysis or degradation of RBPs. A paradigmatic example of this response is observed in virus-infected cells []. Viral encoded proteases produced during picornavirus infection induce the proteolysis of a large number of host factors (Table 1) including splicing factors, RNA-processing proteins, RNA helicases or nuclear pore factors [–], leading to a redistribution of nuclear proteins to the cell cytoplasm. In addition, proteolytic cleavage of eukaryotic initiation factors (eIFs) [–] inhibits protein synthesis and in general, causes a shut-down of cellular gene expression. Specifically, cleavage of eIF4GI and PABP by picornavirus-encoded proteases induces the shut-off of cap-dependent translation in infected cells.

Table 1.
RNA-binding proteins proteolyzed in picornavirus infected cells.
Traditional RBPs consist of modular RNA-binding domains (RBDs), typically RNA recognition motifs (RRM), heterogeneous nuclear ribonucleoprotein K-homology domains (KH), cold-shock domains (CSD), zinc finger domains (ZNF), double-stranded RNA-binding domains (dsRBD), Piwi/Argonaute/Zwile (PAZ) domain, RGG (Arg-Gly-Gly) box, DEAD/DEAH box, Sm domains and Pumilio/FBF (PUF or Pum-HD) domains []. Representative examples of RBPs are the polypyrimidin tract-binding protein (PTB), the hnRNP K, the upstream of N-ras (Unr), or the Zinc finger protein 9 (ZNF9). Among well characterized RBPs are hnRNPs, a large family of nuclear proteins (hnRNP A1 to hnRNP U) with RNA-binding domains and protein-protein binding motifs [] that shuttle with the RNA from the nucleus to the cytoplasm and regulate transcription, splicing, RNA turnover and translation. PTB (also termed hnRNP I) is a protein with four RRMs that recognize U/C-rich sequences []. HnRNP K, PCBP1 (hnRNP E1) and PCBP2 (hnRNP E2) recognize polyr(C) regions and share the KH RNA-binding domain []. Unr is a cold-shock domain RBP that interacts with the poly(A)-binding protein (PABP) []. Zinc finger proteins, however, are DNA and RNA-binding proteins.
Given the many layers of post-transcriptional control operating in the cell, the number of factors that can be involved in various steps of RNA function is much larger than anticipated. Indeed, the recent development of methodologies aimed at searching for new RBPs has produced a catalogue of factors with RNA binding capacity [], such as enzymes of intermediary metabolism among others. In the near future, in depth characterization will indicate whether these factors are passengers of RNP complexes or will provide evidences for the functional role of these factors in RNA-dependent pathways.
2. RNA-Binding Proteins and Translation Control
Most cellular mRNAs initiate translation by a mechanism that depends on the recognition of the m7G(5′)ppp(5′)N structure (termed cap) located at the 5′ end of most mRNAs []. In this mechanism, the 5′ cap structure of the mRNA is recognized by eIF4F, a complex composed of the cap-binding factor eIF4E, the scaffolding protein eIF4G and the RNA helicase eIF4A. The cap-binding capacity of eIF4E is regulated by phosphorylation level of eIF4E-binding proteins (eIF4E-BP 1-3). In turn, eIF4G interacts with eIF3 and PABP, the protein interacting with the poly(A) tail of the mRNA. In addition, the middle region of eIF4G around the first HEAT motif displays RNA binding capacity []. The thirteen multimeric factor eIF3 (eIF3a-eIF3m) is organized as a five-lobed structure []. Several subunits harbor RNA-binding domains (eIF3b and eIF3g) or have being involved in direct binding to mRNA (eIF3d). In addition, recent analysis of the interaction of eIF3 with a viral RNA has identified helix-loop-helix (HLH) motif [] in eIF3a-c as the region responsible for RNA interaction.
On the other hand, the 40S ribosomal subunit with the ternary complex (TC) (consisting of the initiator methionyl-tRNAi and eIF2-GTP) mediates the formation of the 43S complex that is recruited to the mRNA along with eIF1A, eIF1, eIF3, and possibly eIF5. Assembly of a competent 43S complex into mRNA bound to eIF4F is further stabilized by the interaction of eIF4G with PABP, and of eIF4B with eIF4A and PABP. The protein eIF4B harbors an RRM motif and a C-terminal Arginine-rich motif required for RNA-binding []. The 43S complex scans the 5′ UTR region of the mRNA until the first initiation codon in the proper context is encountered, leading to the formation of the 48S initiation complex. At this step, eIF1 ensures fidelity of initiation codon selection, discriminating against non-AUG and AUG codons located in poor context []. Furthermore, scanning of highly structured 5′ UTRs depends on the RNA helicase DHX29 []. Following eIF1 displacement, eIF5 mediates the hydrolysis of eIF2-bound GTP and eIF5B mediates joining of the 60S subunit yielding the 80S ribosome that gives rise to the start of polypeptide synthesis. For recent reviews on translation initiation see [,].
Cap-dependent translation initiation is inhibited under cellular stress conditions, such as viral infection or apoptosis [,,]. Proteolysis of eIF4G and PABP and changes in the phosphorylation levels of eIF4E-binding proteins (eIF4E-BPs) severely compromise cap-dependent translation initiation in picornavirus infected cells []. These adverse situations, however, allow translation of some viral mRNAs and a subset of cellular mRNAs that evade translation shut-down, taking advantage of cis-acting regulatory elements known as internal ribosome entry site (IRES) elements. Translation of mRNAs bearing IRES elements, first reported in the genomic RNA of picornaviruses [,], is therefore resistant to cap-dependent shut-down.
Soon after picornavirus IRES disclosure, other viral RNAs [,] and cellular mRNAs possessing IRES elements were discovered due to their capacity to remain attached to polysomes under conditions that inhibit cap-dependent translation []. Cellular mRNAs bearing IRES elements do contain a cap at the 5′ end although they are translated at low levels, but can switch to an IRES-dependent mechanism when cap-dependent initiation is compromised []. In fact, there are examples where translation is increased (vimentin) and others where translation persists despite shut-down of cap-dependent translation (myc or nucleophosmin) []. The internal initiation process is assisted by RBPs, which are thought to facilitate the proper folding of the IRES region allowing the recruitment of the translation machinery []. The list of mRNAs that can be translated using cap-independent mechanisms is growing constantly [–]. For other IRES reports, see recent reviews [,]. Most IRES elements are located within the 5′ UTR of mRNAs. However, examples of IRES elements located within the coding sequence also exist [–]. In these cases, the polypeptide expressed from the internal initiation codon results in a shorter protein with different function [,]. Thereby, this manner of translation initiation opens new avenues for gene expression control.
Although the presence of IRES elements in viral RNAs is well established, data on some cellular elements has been a matter of debate due to the lack of appropriate controls performed to discard the presence of cryptic promoters or alternative splicing transcripts [,]. Indeed, conserved features of IRES elements in cellular mRNAs remain largely unknown since they differ in nucleotide sequence, RNA secondary structure and IRES trans-acting factors (ITAFs) requirement [–]. Moreover, the lack of conserved features hampers prediction of novel IRES elements using computational methods.
According to the minimal requirement of factors for internal initiation, viral IRES elements can be grouped into two categories. The first category, represented by the intergenic region (IGR) of dicistroviruses, includes those that do not need proteins to assemble the initiation complex. This unique class of IRES elements adopts a compact tertiary structure that functionally substitutes the initiator met-tRNAi during internal initiation, driving protein synthesis without the help of eIFs at non-AUG codons [,,]. The second category includes those elements that do need eIFs and RBPs to recruit the ribosome, such as picornavirus or cellular IRES elements [–]. In addition, distinct groups can be made within the second category, depending on the RNA structural organization and the proteins required for activity (see below).
3. RNA-Binding Proteins Modulating Viral IRES Activity
RNA structure plays a fundamental role in viral IRES-dependent translation initiation [–]. Structural and functional studies have shown that RNA structure of viral IRES elements is organized in phylogenetically conserved modules [,–], suggesting a distribution of functions among the different domains of an IRES element [,]. Evidence for links between RNA structure and biological function is also supported by the conservation of structural motifs within IRES elements of highly variable genomes [–]. For the sake of brevity, we will discuss in this review the IRES elements located in the genome of picornavirus and hepacivirus.
3.2. RNA-Binding Proteins Modulating Hepatitis C IRES Activity
The IRES element of HCV genome differs from picornavirus IRES elements in RNA structure and eIF4G requirement [,]. The HCV IRES is organized in three structural domains (designated II, III and IV) [], although destabilization of domain IV is not detrimental to IRES function []. In the absence of eIFs, domain III can form a high-affinity complex with the 40S ribosomal subunit []. However, eIF3 and eIF2 are necessary for 48S initiation complex formation in vitro [] despite the fact that HCV IRES activity has been shown to be partially resistant to eIF2 inactivation []. Recent cryoEM studies have contributed to the understanding of the interaction of eIF3 with the HCV IRES []. Mutations in the RNA-binding motif of eIF3a weaken eIF3 binding to the HCV IRES and the 40S ribosomal subunit, suppressing eIF2-dependent recognition of the start codon. Mutations in the eIF3c RNA-binding motif also reduce 40S ribosomal subunit binding to eIF3 and inhibit eIF5B-dependent steps downstream of start codon recognition.
In addition to eIF3 and the ternary complex, a few RBPs acting as ITAFs are shared between HCV and picornavirus IRES elements (PTB, PCBP2, Nucleolin, Gemin5, Unr, hnRNPA1/A2, La autoantigen (La) and NS1-associated protein (NSAP1, also known as hnRNP D) [,,–]). Whether these RBPs modulate translation initiation promoted by other viral IRES remains to be elucidated.
Both picornavirus and HCV IRES-dependent translation are synergistically enhanced by the 3′ UTR of the viral genome [–], consistent with a functional link between the 5′ and 3′ ends of the viral RNA. In picornavirus RNAs, the 3′ UTR is composed of two stem-loops and a short poly(A) tail that are required for virus multiplication. In contrast, the HCV viral RNA possesses a poly(U) tract and a complex RNA structure located near the 3′ end. Bridging 5′ and 3′ ends of viral RNAs involves direct RNA-RNA contacts and RNA-protein interactions [,]. Accordingly, riboproteomic procedures on RNAs with two distant cis-acting regions identified factors mediating the formation of functional bridges between mRNA regions. This was the case of RNPs assembled on tagged RNAs that contained both the IRES and the 3′ UTR of HCV []. One of the identified proteins, the insulin-like growth factor II mRNA-binding protein 1 (IGF2BP1), coimmunoprecipitates with eIF3 and the 40S ribosomal subunit, suggesting that it enhances HCV IRES-dependent translation by recruiting the ribosomal subunits to a pseudo-circularized RNA. Recent studies have proposed that 3′ UTR interaction with the ribosomal subunit retains ribosome complexes during translation termination, facilitating efficient initiation of subsequent rounds of translation [].
4. RNA-Binding Proteins Controlling Cellular IRES Activity
Cellular IRES elements are typically present in mRNAs encoding stress response proteins, such as those needed during nutrient deprivation, temperature shock, hibernation, hypoxia, cell cycle arrest, or apoptosis [,]. Hence, they have evolved mechanisms to evade global repression of translation. The study of some cellular IRES elements has shown that ITAFs facilitate the binding of the mRNA to ribosomal 40S subunits, in conjunction with eIF2, eIF3, eIF4F, and PABP [–]. Accordingly, changes in ITAFs abundance, post-translational modifications or subcellular localization, modulate internal initiation of translation during different stress conditions [].
Multifunctional RBPs interact with various IRES elements, suggesting a mechanism for the coordinated regulation of translation initiation of a subset of mRNAs bound by shuttling proteins, such as hnRNPs and splicing factors (Table 3). Nuclear proteins found in complexes associated with various gene expression steps involving RNA molecules (PTB, the splicing-factor related protein proline and glutamine-rich SFPQ/PSF, the non-POU domain-containing octamer binding nuclear RNA binding protein (nonO/p54nrb), PCBP2, or HuR) control the expression of lymphoid enhancer binding factor (LEF1), c-myc, CDK11, or p53 [,–]. However, there are other cases where specific RBPs (such as mouse hnRNP Q and FMRP) are reported to control a reduced number of IRES elements [,]. This differential regulation suggests that specialized RBPs might exert their function in translation control by binding to the IRES region of specific cellular mRNAs during splicing complex assembly before nuclear export. It remains to be determined whether this is the result of a specialized activity or if it reflects the lack of sufficient detailed studies. Below, we discuss the capacity of various RBPs to control cellular IRES elements driving the expression of proteins grouped according to their function in the cell. In this process, deleting the content is not recommended.

Table 3.
RNA-binding proteins m odulating cellular IRES.
4.1. ITAFs Controlling the Expression of Cell Proliferation Proteins
PTB stimulates translation driven by IRES elements located in mRNAs encoding proteins of the myc family controlling cell growth, the tumor suppressor gene p53 as well as other factors involved in apoptosis and nutrient deprivation [,,]. A complex formed by Annexin A2, PSF and PTB binds and stimulates p53 IRES in the presence of calcium ions []. The unr, c-myc, CDK11, and serine/threonine-protein kinase PITSLREp58 IRES elements are activated during mitosis [,], a cell cycle phase where cap-dependent translation is compromised. Protein-protein interaction and/or coordinated RNA-proteins complex assembly influence internal initiation, as shown in the case of IRES activity of c-myc and PITSLRE mRNAs, whose function depends on the Unr-partners, hnRNP K, PCBP1-2, or hnRNP C1-2, respectively [,]. On the other hand, stress-dependent modifications or relocalization of hnRNP A1 mediates internal initiation of c-myc, unr, cyclin D1, or sterol-regulatory-element-binding protein 1 (SREBP-1a) mRNAs [,].
Translation of specific mRNAs in cells with quiescent v-akt murine thymoma viral oncogene homolog 1 (AKT) kinase maintains the levels of proteins involved in cell cycle progression when eIF4E-mediated (cap-dependent) translation is inhibited. This pathway is dependent on SAPK2/p38-mediated activation of IRES-dependent initiation of the cyclin D1 and c-myc mRNAs []. Inhibition of SAPK2/p38 in glioblastoma multiforme cells reduces rapamycin-induced IRES-mediated translation initiation of cyclin D1 and c-myc, resulting in G1 arrest and inhibition of tumor growth.
4.2. ITAFs Controlling Translation of Pro-apoptotic and Pro-survival mRNAs
IRES located in mRNAs encoding proteins synthesized under apoptosis such as the apoptotic protease activating factor 1 (Apaf-1), and BCL2-associated athanogene (BAG-1), are also responsive to PTB []. In particular, IRES activity of Apaf-1 mRNA is regulated via PTB and Unr []. However, during apoptosis the Apaf-1 IRES is activated while the X-linked inhibitor of apoptosis protein (XIAP) is inhibited []. It has been reported that relocalization of hnRNP A1 mediates internal initiation of Apaf-1 and XIAP [,]. Other proteins such as DAP5 and HuR exert a stimulatory role on apoptotic mRNAs [,].
With the exception of pyrimidine tracts, no distinctive RNA motifs that can be used to predict the binding of RBPs are apparent in cellular IRES elements. Yet, cellular IRES with high AU content, such as XIAP, depend on NF45 [], since cells deficient in NF45 exhibit reduced IRES-mediated translation of XIAP and cellular inhibitor of apoptosis protein 1 (cIAP1) mRNAs that, in turn, leads to dysregulated expression of survivin and cyclin E.
Although most ITAFs stimulate translation, a few cases where RBPs repress translation have also been reported. IRES-dependent translation of anti-apoptotic proteins XIAP and Bcl-X is repressed by the tumor suppressor programmed cell death 4 (PDCD4), a factor that sequesters eIF4A and, thus, inhibits formation of the 48S translation initiation complex []. Phosphorylation of PDCD4 by activated S6K2 leads to the degradation of PDCD4, stimulating XIAP and Bcl-x(L) translation.
4.3. ITAFs Controlling Response to Nutrient Starvation, ER Stress, or Growth Factors
As in the case of IRES elements controlling the expression of cell proliferation proteins, stress-dependent relocalization of hnRNP A1 also mediates internal initiation of vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF-2) []. However, a negative effect of DKC1 on the VEGF IRES has been recently reported [], likely due to a reduction on ribosome availability. HuR was found to negatively affect translation of the IGF1 receptor or the thrombomodulin (TM) endothelial cell receptor [] by yet unknown mechanisms. A different case of a repressor ITAF is the DEAD-box RNA helicase 6 (DDX6) []. DDX6 inhibits VEGF IRES-mediated translation in normoxic MCF-7 extract. However, under hypoxia the level of DDX6 declines and its interaction with VEGF mRNA is diminished in vivo. In addition, DDX6 knockdown cells show increased secretion of VEGF.
The death-associated protein (DAP5), NF45, G-rich RNA sequence binding factor (GRSF-1), fragile-X mental retardation protein (FMRP), heterogeneous nuclear ribonucleoprotein D-like protein (JKTBP1) and zinc-finger protein ZNF9 are specific for some IRES elements such as DAP5/p97, cIAP1, ODC [,,,]. Moreover, circadian expression of mouse PERIOD1 (mPER1) is regulated by rhythmic translational control of mPer1 mRNA, together with transcriptional modulation, via an IRES element along with the mouse trans-acting factor mhnRNP Q []. The rate of IRES-mediated translation exhibits phase-dependent characteristics through rhythmic interactions between mPer1 mRNA and mhnRNP Q.
In summary, the data available so far indicate that individual mRNAs exploit the activity of multifunctional RBPs to overcome the global repression of protein synthesis.
5. Concluding Remarks and Perspectives
ITAFs are RBPs that activate or repress the expression of proteins critical in the cellular response to growth, nutritional, environmental and proliferation signals. It has been proposed that the main role of RBPs is to remodel the mRNA structure in a way that enhances its affinity for components of the translation machinery, or that they substitute some canonical eIFs providing functional bridges between the mRNA and the ribosomal subunits. Elucidating the function of ITAFs demands a deep understanding of their RNA targets and their protein partners with potential modifications. Yet, a subset of IRES elements exhibiting different structural organization interacts with the ribosomal protein RpS25 [], consistent with the observation that two of these IRES elements (HCV and the dicistrovirus IGR) induce similar conformational changes in the 40S ribosomal subunit despite their different RNA structural organization []. Indeed, the IRES property of interacting with the ribosomal RNA [,] is an attractive idea that is also consistent with the finding that IRES activity is sensitive to changes in the ribosome composition [–]. In the near future, the characterization at the molecular level of more IRES elements will shed new light on the understanding of the strategies used to recruit the translation machinery. In turn, this will open new avenues to predict novel regulatory elements using this specialized mechanism of translation initiation.
Acknowledgments
This work was supported by grants BFU2011-25437, CSD2009-00080 and by an institutional grant from Fundación Ramón Areces.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Muller-McNicoll, M.; Neugebauer, K.M. How cells get the message: Dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet 2013, 14, 275–287. [Google Scholar]
- Cooper, T.A.; Wan, L.; Dreyfuss, G. RNA and disease. Cell 2009, 136, 777–793. [Google Scholar]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar]
- Walsh, D.; Mohr, I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol 2011, 9, 860–875. [Google Scholar]
- Spriggs, K.A.; Bushell, M.; Willis, A.E. Translational regulation of gene expression during conditions of cell stress. Mol. Cell 2010, 40, 228–237. [Google Scholar]
- Liwak, U.; Faye, M.D.; Holcik, M. Translation control in apoptosis. Exp. Oncol 2012, 34, 218–230. [Google Scholar]
- Komar, A.A.; Hatzoglou, M. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 2011, 10, 229–240. [Google Scholar]
- Almstead, L.L.; Sarnow, P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes Dev 2007, 21, 1086–1097. [Google Scholar]
- Mukherjee, A.; Morosky, S.A.; Delorme-Axford, E.; Dybdahl-Sissoko, N.; Oberste, M.S.; Wang, T.; Coyne, C.B. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 2011, 7, e1001311. [Google Scholar]
- Perera, R.; Daijogo, S.; Walter, B.L.; Nguyen, J.H.; Semler, B.L. Cellular protein modification by poliovirus: The two faces of poly(rC)-binding protein. J. Virol 2007, 81, 8919–8932. [Google Scholar]
- Barral, P.M.; Sarkar, D.; Fisher, P.B.; Racaniello, V.R. RIG-I is cleaved during picornavirus infection. Virology 2009, 391, 171–176. [Google Scholar]
- Weng, K.F.; Li, M.L.; Hung, C.T.; Shih, S.R. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog 2009, 5, e1000593. [Google Scholar]
- Chen, L.L.; Kung, Y.A.; Weng, K.F.; Lin, J.Y.; Horng, J.T.; Shih, S.R. Enterovirus 71 infection cleaves a negative regulator for viral internal ribosomal entry site-driven translation. J. Virol 2013, 87, 3828–3838. [Google Scholar]
- Castello, A.; Izquierdo, J.M.; Welnowska, E.; Carrasco, L. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J. Cell Sci 2009, 122, 3799–3809. [Google Scholar] [Green Version]
- Shiroki, K.; Isoyama, T.; Kuge, S.; Ishii, T.; Ohmi, S.; Hata, S.; Suzuki, K.; Takasaki, Y.; Nomoto, A. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol 1999, 73, 2193–2200. [Google Scholar]
- Rozovics, J.M.; Chase, A.J.; Cathcart, A.L.; Chou, W.; Gershon, P.D.; Palusa, S.; Wilusz, J.; Semler, B.L. Picornavirus modification of a host mRNA decay protein. mBio 2012, 3, e00431–e00412. [Google Scholar]
- Watters, K.; Palmenberg, A.C. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J. Virol 2011, 85, 10874–10883. [Google Scholar]
- Pineiro, D.; Ramajo, J.; Bradrick, S.S.; Martinez-Salas, E. Gemin5 proteolysis reveals a novel motif to identify L protease targets. Nucleic Acids Res 2012, 40, 4942–4953. [Google Scholar]
- White, J.P.; Cardenas, A.M.; Marissen, W.E.; Lloyd, R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2007, 2, 295–305. [Google Scholar]
- Lawrence, P.; Schafer, E.A.; Rieder, E. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 2012, 425, 40–52. [Google Scholar]
- Park, N.; Skern, T.; Gustin, K.E. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J. Biol. Chem 2010, 285, 28796–28805. [Google Scholar]
- Belsham, G.J.; McInerney, G.M.; Ross-Smith, N. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J. Virol 2000, 74, 272–280. [Google Scholar]
- De Breyne, S.; Bonderoff, J.M.; Chumakov, K.M.; Lloyd, R.E.; Hellen, C.U. Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology 2008, 378, 118–122. [Google Scholar]
- Rodriguez Pulido, M.; Serrano, P.; Saiz, M.; Martinez-Salas, E. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology 2007, 364, 466–474. [Google Scholar]
- Bonderoff, J.M.; Larey, J.L.; Lloyd, R.E. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation. J. Virol 2008, 82, 9389–9399. [Google Scholar]
- Etchison, D.; Milburn, S.C.; Edery, I.; Sonenberg, N.; Hershey, J.W.B. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000 dalton polypeptide associated with eukaryotic initiation factor 3 and a cap-binding complex. J. Biol. Chem 1982, 257, 14806–14810. [Google Scholar]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008, 582, 1977–1986. [Google Scholar]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol 2007, 8, 479–490. [Google Scholar]
- Conte, M.R.; Grune, T.; Ghuman, J.; Kelly, G.; Ladas, A.; Matthews, S.; Curry, S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J 2000, 19, 3132–3141. [Google Scholar]
- Makeyev, A.V.; Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002, 8, 265–278. [Google Scholar]
- Chang, T.C.; Yamashita, A.; Chen, C.Y.; Yamashita, Y.; Zhu, W.; Durdan, S.; Kahvejian, A.; Sonenberg, N.; Shyu, A.B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev 2004, 18, 2010–2023. [Google Scholar]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar]
- Marcotrigiano, J.; Lomakin, I.B.; Sonenberg, N.; Pestova, T.V.; Hellen, C.U.; Burley, S.K. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 2001, 7, 193–203. [Google Scholar]
- Siridechadilok, B.; Fraser, C.S.; Hall, R.J.; Doudna, J.A.; Nogales, E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 2005, 310, 1513–1515. [Google Scholar]
- Sun, C.; Querol-Audi, J.; Mortimer, S.A.; Arias-Palomo, E.; Doudna, J.A.; Nogales, E.; Cate, J.H. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res 2013, 41, 7512–7521. [Google Scholar]
- Walker, S.E.; Zhou, F.; Mitchell, S.F.; Larson, V.S.; Valasek, L.; Hinnebusch, A.G.; Lorsch, J.R. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA 2013, 19, 191–207. [Google Scholar]
- Lomakin, I.B.; Shirokikh, N.E.; Yusupov, M.M.; Hellen, C.U.; Pestova, T.V. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J 2006, 25, 196–210. [Google Scholar]
- Pisareva, V.P.; Pisarev, A.V.; Komar, A.A.; Hellen, C.U.; Pestova, T.V. Translation initiation on mammalian mRNAs with structured 5′ UTRs requires DExH-box protein DHX29. Cell 2008, 135, 1237–1250. [Google Scholar]
- Martinez-Salas, E.; Pacheco, A.; Serrano, P.; Fernandez, N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol 2008, 89, 611–626. [Google Scholar]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1998, 334, 320–325. [Google Scholar]
- Jang, S.K.; Krausslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol 1998, 62, 2636–2643. [Google Scholar]
- Honda, M.; Ping, L.H.; Rijnbrand, R.C.; Amphlett, E.; Clarke, B.; Rowlands, D.; Lemon, S.M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology 1996, 222, 31–42. [Google Scholar]
- Wilson, J.E.; Pestova, T.V.; Hellen, C.U.; Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell 2000, 102, 511–520. [Google Scholar]
- Macejak, D.G.; Sarnow, P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature 1991, 353, 90–94. [Google Scholar]
- Johannes, G.; Sarnow, P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 1998, 4, 1500–1513. [Google Scholar]
- Qin, X.; Sarnow, P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem 2004, 279, 13721–13728. [Google Scholar]
- Spriggs, K.A.; Bushell, M.; Mitchell, S.A.; Willis, A.E. Internal ribosome entry segment-mediated translation during apoptosis: The role of IRES-trans-acting factors. Cell Death Differ 2005, 12, 585–591. [Google Scholar]
- Spriggs, K.A.; Cobbold, L.C.; Ridley, S.H.; Coldwell, M.; Bottley, A.; Bushell, M.; Willis, A.E.; Siddle, K. The human insulin receptor mRNA contains a functional internal ribosome entry segment. Nucleic Acids Res 2009, 37, 5881–5893. [Google Scholar]
- Villa-Cuesta, E.; Sage, B.T.; Tatar, M. A role for Drosophila dFoxO and dFoxO 5′ UTR internal ribosomal entry sites during fasting. PLoS One 2010, 5, e11521. [Google Scholar]
- Marr, M.T.; D’Alessio, J.A., 2nd; Puig, O.; Tjian, R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev 2007, 21, 175–183. [Google Scholar]
- Dinkova, T.D.; Zepeda, H.; Martinez-Salas, E.; Martinez, L.M.; Nieto-Sotelo, J.; de Jimenez, E.S. Cap-independent translation of maize Hsp101. Plant J 2005, 41, 722–731. [Google Scholar]
- Du, X.; Wang, J.; Zhu, H.; Rinaldo, L.; Lamar, K.M.; Palmenberg, A.C.; Hansel, C.; Gomez, C.M. Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell 2013, 154, 118–133. [Google Scholar]
- Al-Fageeh, M.B.; Smales, C.M. Cold-inducible RNA binding protein (CIRP) expression is modulated by alternative mRNAs. RNA 2009, 15, 1164–1176. [Google Scholar]
- Fox, J.T.; Shin, W.K.; Caudill, M.A.; Stover, P.J. A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair. J. Biol. Chem 2009, 284, 31097–31108. [Google Scholar]
- Rubsamen, D.; Blees, J.S.; Schulz, K.; Doring, C.; Hansmann, M.L.; Heide, H.; Weigert, A.; Schmid, T.; Brune, B. IRES-dependent translation of egr2 is induced under inflammatory conditions. RNA 2012, 18, 1910–1920. [Google Scholar]
- Timmerman, S.L.; Pfingsten, J.S.; Kieft, J.S.; Krushel, L.A. The 5′ leader of the mRNA encoding the mouse neurotrophin receptor TrkB contains two internal ribosomal entry sites that are differentially regulated. PLoS One 2008, 3, e3242. [Google Scholar]
- Yin, J.Y.; Dong, Z.Z.; Liu, R.Y.; Chen, J.; Liu, Z.Q.; Zhang, J.T. Translational regulation of RPA2 via internal ribosomal entry site and by eIF3a. Carcinogenesis 2013, 34, 1224–1231. [Google Scholar]
- Blau, L.; Knirsh, R.; Ben-Dror, I.; Oren, S.; Kuphal, S.; Hau, P.; Proescholdt, M.; Bosserhoff, A.K.; Vardimon, L. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc. Natl. Acad. Sci. USA 2012, 109, E2875–E2884. [Google Scholar]
- Veo, B.L.; Krushel, L.A. Secondary RNA structure and nucleotide specificity contribute to internal initiation mediated by the human tau 5′ leader. RNA Biol 2012, 9, 1344–1360. [Google Scholar]
- Martinez-Salas, E.; Pineiro, D.; Fernandez, N. Alternative mechanisms to initiate translation in eukaryotic mRNAs. Comp. Funct. Genomics 2012, 2012, 391546:1–391546:12. [Google Scholar]
- Le, H.T.; Sorrell, A.M.; Siddle, K. Two isoforms of the mRNA binding protein IGF2BP2 are generated by alternative translational initiation. PLoS One 2012, 7, e33140. [Google Scholar]
- Weingarten-Gabbay, S.; Khan, D.; Liberman, N.; Yoffe, Y.; Bialik, S.; Das, S.; Oren, M.; Kimchi, A. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 2013. [Google Scholar] [CrossRef]
- Locker, N.; Chamond, N.; Sargueil, B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res 2011, 39, 2367–2377. [Google Scholar]
- Burkart, C.; Fan, J.B.; Zhang, D.E. Two independent mechanisms promote expression of an N-terminal truncated USP18 isoform with higher DeISGylation activity in the nucleus. J. Biol. Chem 2012, 287, 4883–4893. [Google Scholar]
- Bert, A.G.; Grepin, R.; Vadas, M.A.; Goodall, G.J. Assessing IRES activity in the HIF-1α and other cellular 5′ UTRs. RNA 2006, 12, 1074–1083. [Google Scholar]
- Baranick, B.T.; Lemp, N.A.; Nagashima, J.; Hiraoka, K.; Kasahara, N.; Logg, C.R. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl. Acad. Sci. USA 2008, 105, 4733–4738. [Google Scholar]
- Fernandez, J.; Yaman, I.; Huang, C.; Liu, H.; Lopez, A.B.; Komar, A.A.; Caprara, M.G.; Merrick, W.C.; Snider, M.D.; Kaufman, R.J.; et al. Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol. Cell 2005, 17, 405–416. [Google Scholar]
- Le Quesne, J.P.; Stoneley, M.; Fraser, G.A.; Willis, A.E. Derivation of a structural model for the c-myc IRES. J. Mol. Biol 2001, 310, 111–126. [Google Scholar]
- Morris, M.J.; Negishi, Y.; Pazsint, C.; Schonhoft, J.D.; Basu, S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc 2010, 132, 17831–17839. [Google Scholar]
- Sasaki, J.; Nakashima, N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999, 73, 1219–1226. [Google Scholar]
- Costantino, D.A.; Pfingsten, J.S.; Rambo, R.P.; Kieft, J.S. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol 2008, 15, 57–64. [Google Scholar]
- Pestova, T.V.; Shatsky, I.N.; Fletcher, S.P.; Jackson, R.J.; Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 1998, 12, 67–83. [Google Scholar]
- Lopez de Quinto, S.; Martinez-Salas, E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 2000, 6, 1380–1392. [Google Scholar]
- Mitchell, S.A.; Spriggs, K.A.; Coldwell, M.J.; Jackson, R.J.; Willis, A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 2003, 11, 757–771. [Google Scholar]
- Filbin, M.E.; Kieft, J.S. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol 2009, 19, 267–276. [Google Scholar]
- Martinez-Salas, E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol 2008, 16, 230–237. [Google Scholar]
- Perard, J.; Leyrat, C.; Baudin, F.; Drouet, E.; Jamin, M. Structure of the full-length HCV IRES in solution. Nat. Commun 2013. [Google Scholar] [CrossRef]
- Ren, Q.; Wang, Q.S.; Firth, A.E.; Chan, M.M.; Gouw, J.W.; Guarna, M.M.; Foster, L.J.; Atkins, J.F.; Jan, E. Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site. Proc. Natl. Acad. Sci. USA 2012, 109, E630–E639. [Google Scholar]
- Fernandez, N.; Garcia-Sacristan, A.; Ramajo, J.; Briones, C.; Martinez-Salas, E. Structural analysis provides insights into the modular organization of picornavirus IRES. Virology 2011, 409, 251–261. [Google Scholar]
- Fernandez, N.; Buddrus, L.; Pineiro, D.; Martinez-Salas, E. Evolutionary conserved motifs constrain the RNA structure organization of picornavirus IRES. FEBS Lett 2013, 587, 1353–1358. [Google Scholar]
- Serrano, P.; Ramajo, J.; Martinez-Salas, E. Rescue of internal initiation of translation by RNA complementation provides evidence for a distribution of functions between individual IRES domains. Virology 2009, 388, 221–229. [Google Scholar]
- Jang, C.J.; Jan, E. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA 2010, 16, 1182–1195. [Google Scholar]
- Barria, M.I.; Gonzalez, A.; Vera-Otarola, J.; Leon, U.; Vollrath, V.; Marsac, D.; Monasterio, O.; Perez-Acle, T.; Soza, A.; Lopez-Lastra, M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res 2009, 37, 957–971. [Google Scholar]
- Fajardo, T., Jr.; Rosas, M.F.; Sobrino, F.; Martinez-Salas, E. Exploring IRES region accessibility by Interference of foot-and-mouth disease virus infectivity. PLoS One 2012, 7, e41382. [Google Scholar]
- Fernandez-Miragall, O.; Ramos, R.; Ramajo, J.; Martinez-Salas, E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA 2006, 12, 223–234. [Google Scholar]
- Fernandez, N.; Fernandez-Miragall, O.; Ramajo, J.; Garcia-Sacristan, A.; Bellora, N.; Eyras, E.; Briones, C.; Martinez-Salas, E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res 2011, 39, 8572–8585. [Google Scholar]
- Fernandez-Miragall, O.; Lopez de Quinto, S.; Martinez-Salas, E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res 2009, 139, 172–182. [Google Scholar]
- Kolupaeva, V.G.; Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem 1998, 273, 18599–18604. [Google Scholar]
- De Breyne, S.; Yu, Y.; Unbehaun, A.; Pestova, T.V.; Hellen, C.U. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. USA 2009, 106, 9197–9202. [Google Scholar]
- Lopez de Quinto, S.; Lafuente, E.; Martinez-Salas, E. IRES interaction with translation initiation factors: Functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 2001, 7, 1213–1226. [Google Scholar]
- Pestova, T.V.; Shatsky, I.N.; Hellen, C.U. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell Biol 1996, 16, 6870–6878. [Google Scholar]
- Andreev, D.E.; Fernandez-Miragall, O.; Ramajo, J.; Dmitriev, S.E.; Terenin, I.M.; Martinez-Salas, E.; Shatsky, I.N. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA 2007, 13, 1366–1374. [Google Scholar]
- Lopez de Quinto, S.; Martinez-Salas, E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology 1999, 255, 324–336. [Google Scholar]
- Redondo, N.; Sanz, M.A.; Steinberger, J.; Skern, T.; Kusov, Y.; Carrasco, L. Translation directed by hepatitis A virus IRES in the absence of active eIF4F complex and eIF2. PLoS One 2012, 7, e52065. [Google Scholar]
- Belsham, G.J. Divergent picornavirus IRES elements. Virus Res 2009, 139, 183–192. [Google Scholar]
- Pisarev, A.V.; Chard, L.S.; Kaku, Y.; Johns, H.L.; Shatsky, I.N.; Belsham, G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol 2004, 78, 4487–4497. [Google Scholar]
- Yu, Y.; Sweeney, T.R.; Kafasla, P.; Jackson, R.J.; Pestova, T.V.; Hellen, C.U. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J 2011, 30, 4423–4436. [Google Scholar]
- Pilipenko, E.V.; Pestova, T.V.; Kolupaeva, V.G.; Khitrina, E.V.; Poperechnaya, A.N.; Agol, V.I.; Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev 2000, 14, 2028–2045. [Google Scholar]
- Jang, S.K.; Wimmer, E. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev 1990, 4, 1560–1572. [Google Scholar]
- Luz, N.; Beck, E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol 1991, 65, 6486–6494. [Google Scholar]
- Gosert, R.; Chang, K.H.; Rijnbrand, R.; Yi, M.; Sangar, D.V.; Lemon, S.M. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites In vivo. Mol. Cell Biol 2000, 20, 1583–1595. [Google Scholar]
- Pacheco, A.; Reigadas, S.; Martinez-Salas, E. Riboproteomic analysis of polypeptides interacting with the internal ribosome-entry site element of foot-and-mouth disease viral RNA. Proteomics 2008, 8, 4782–4790. [Google Scholar]
- Monie, T.P.; Perrin, A.J.; Birtley, J.R.; Sweeney, T.R.; Karakasiliotis, I.; Chaudhry, Y.; Roberts, L.O.; Matthews, S.; Goodfellow, I.G.; Curry, S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J 2007, 26, 3936–3944. [Google Scholar] [Green Version]
- Sean, P.; Nguyen, J.H.; Semler, B.L. Altered interactions between stem-loop IV within the 5′ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: Effects on IRES-mediated translation and viral infectivity. Virology 2009, 389, 45–58. [Google Scholar]
- Fitzgerald, K.D.; Semler, B.L. Re-localization of cellular protein SRp20 during poliovirus infection: Bridging a viral IRES to the host cell translation apparatus. PLoS Pathog 2011, 7, e1002127. [Google Scholar]
- Lin, J.Y.; Li, M.L.; Shih, S.R. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res 2009, 37, 47–59. [Google Scholar]
- Pacheco, A.; Lopez de Quinto, S.; Ramajo, J.; Fernandez, N.; Martinez-Salas, E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res 2009, 37, 582–590. [Google Scholar]
- Andreev, D.E.; Hirnet, J.; Terenin, I.M.; Dmitriev, S.E.; Niepmann, M.; Shatsky, I.N. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res 2012, 40, 5602–5614. [Google Scholar]
- Boussadia, O.; Niepmann, M.; Creancier, L.; Prats, A.C.; Dautry, F.; Jacquemin-Sablon, H. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol 2003, 77, 3353–3359. [Google Scholar]
- Merrill, M.K.; Dobrikova, E.Y.; Gromeier, M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol 2006, 80, 3147–3156. [Google Scholar]
- Weinlich, S.; Huttelmaier, S.; Schierhorn, A.; Behrens, S.E.; Ostareck-Lederer, A.; Ostareck, D.H. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3′ UTR. RNA 2009, 15, 1528–1542. [Google Scholar]
- Martino, L.; Pennell, S.; Kelly, G.; Bui, T.T.; Kotik-Kogan, O.; Smerdon, S.J.; Drake, A.F.; Curry, S.; Conte, M.R. Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res 2012, 40, 1381–1394. [Google Scholar]
- Park, S.M.; Paek, K.Y.; Hong, K.Y.; Jang, C.J.; Cho, S.; Park, J.H.; Kim, J.H.; Jan, E.; Jang, S.K. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res 2011, 39, 7791–7802. [Google Scholar]
- Hahm, B.; Kim, Y.K.; Kim, J.H.; Kim, T.Y.; Jang, S.K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol 1998, 72, 8782–8788. [Google Scholar]
- Pilipenko, E.V.; Viktorova, E.G.; Guest, S.T.; Agol, V.I.; Roos, R.P. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J 2001, 20, 6899–6908. [Google Scholar]
- Kafasla, P.; Morgner, N.; Robinson, C.V.; Jackson, R.J. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J 2010, 29, 3710–3722. [Google Scholar]
- Yu, Y.; Abaeva, I.S.; Marintchev, A.; Pestova, T.V.; Hellen, C.U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res 2011, 39, 4851–4865. [Google Scholar]
- Kafasla, P.; Morgner, N.; Poyry, T.A.; Curry, S.; Robinson, C.V.; Jackson, R.J. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol. Cell 2009, 34, 556–568. [Google Scholar]
- Yu, Y.; Ji, H.; Doudna, J.A.; Leary, J.A. Mass spectrometric analysis of the human 40S ribosomal subunit: Native and HCV IRES-bound complexes. Protein Sci 2005, 14, 1438–1446. [Google Scholar]
- Battle, D.J.; Lau, C.K.; Wan, L.; Deng, H.; Lotti, F.; Dreyfuss, G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 2006, 23, 273–279. [Google Scholar]
- Pineiro, D.; Fernandez, N.; Ramajo, J.; Martinez-Salas, E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res 2013, 41, 1017–1028. [Google Scholar]
- Percipalle, P.; Jonsson, A.; Nashchekin, D.; Karlsson, C.; Bergman, T.; Guialis, A.; Daneholt, B. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res 2002, 30, 1725–1734. [Google Scholar]
- Pineiro, D.; Martinez-Salas, E. RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis. Viruses 2012, 4, 2233–2250. [Google Scholar]
- Otto, G.A.; Puglisi, J.D. The pathway of HCV IRES-mediated translation initiation. Cell 2004, 119, 369–380. [Google Scholar]
- Kieft, J.S.; Zhou, K.; Jubin, R.; Doudna, J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 2001, 7, 194–206. [Google Scholar]
- Terenin, I.M.; Dmitriev, S.E.; Andreev, D.E.; Shatsky, I.N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol 2008, 15, 836–841. [Google Scholar]
- Lopez de Quinto, S.; Saiz, M.; de la Morena, D.; Sobrino, F.; Martinez-Salas, E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res 2002, 30, 4398–4405. [Google Scholar]
- Dobrikova, E.; Florez, P.; Bradrick, S.; Gromeier, M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. USA 2003, 100, 15125–15130. [Google Scholar]
- Song, Y.; Friebe, P.; Tzima, E.; Junemann, C.; Bartenschlager, R.; Niepmann, M. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol 2006, 80, 11579–11588. [Google Scholar]
- Serrano, P.; Pulido, M.R.; Saiz, M.; Martinez-Salas, E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol 2006, 87, 3013–3022. [Google Scholar]
- Romero-Lopez, C.; Berzal-Herranz, A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 2009, 15, 1740–1752. [Google Scholar]
- Bai, Y.; Zhou, K.; Doudna, J.A. Hepatitis C virus 3′ UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res 2013, 41, 7861–7874. [Google Scholar]
- Spriggs, K.A.; Cobbold, L.C.; Jopling, C.L.; Cooper, R.E.; Wilson, L.A.; Stoneley, M.; Coldwell, M.J.; Poncet, D.; Shen, Y.C.; Morley, S.J.; et al. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol. Cell Biol 2009, 29, 1565–1574. [Google Scholar]
- Majumder, M.; Yaman, I.; Gaccioli, F.; Zeenko, V.V.; Wang, C.; Caprara, M.G.; Venema, R.C.; Komar, A.A.; Snider, M.D.; Hatzoglou, M. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol. Cell Biol 2009, 29, 2899–2912. [Google Scholar]
- Cobbold, L.C.; Spriggs, K.A.; Haines, S.J.; Dobbyn, H.C.; Hayes, C.; de Moor, C.H.; Lilley, K.S.; Bushell, M.; Willis, A.E. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell Biol 2008, 28, 40–49. [Google Scholar]
- Bushell, M.; Stoneley, M.; Kong, Y.W.; Hamilton, T.L.; Spriggs, K.A.; Dobbyn, H.C.; Qin, X.; Sarnow, P.; Willis, A.E. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell 2006, 23, 401–412. [Google Scholar]
- Lewis, S.M.; Holcik, M.; For, IRES. trans-acting factors, it is all about location. Oncogene 2008, 27, 1033–1035. [Google Scholar]
- Tsai, B.P.; Wang, X.; Huang, L.; Waterman, M.L. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteomics 2011. [Google Scholar] [CrossRef]
- Sharathchandra, A.; Lal, R.; Khan, D.; Das, S. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol 2012, 9, 1429–1439. [Google Scholar]
- Ohno, S.; Shibayama, M.; Sato, M.; Tokunaga, A.; Yoshida, N. Polypyrimidine tract-binding protein regulates the cell cycle through IRES-dependent translation of CDK11(p58) in mouse embryonic stem cells. Cell Cycle 2011, 10, 3706–3713. [Google Scholar]
- Evans, J.R.; Mitchell, S.A.; Spriggs, K.A.; Ostrowski, J.; Bomsztyk, K.; Ostarek, D.; Willis, A.E. Members of the poly(rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene 2003, 22, 8012–8020. [Google Scholar]
- Bechara, E.G.; Didiot, M.C.; Melko, M.; Davidovic, L.; Bensaid, M.; Martin, P.; Castets, M.; Pognonec, P.; Khandjian, E.W.; Moine, H.; et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol 2009, 7, e16. [Google Scholar]
- Lee, K.H.; Woo, K.C.; Kim, D.Y.; Kim, T.D.; Shin, J.; Park, S.M.; Jang, S.K.; Kim, K.T. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol 2012, 32, 717–728. [Google Scholar]
- Grover, R.; Ray, P.S.; Das, S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle 2008, 7, 2189–2198. [Google Scholar]
- Dobbyn, H.C.; Hill, K.; Hamilton, T.L.; Spriggs, K.A.; Pickering, B.M.; Coldwell, M.J.; de Moor, C.H.; Bushell, M.; Willis, A.E. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene 2008, 27, 1167–1174. [Google Scholar]
- Tinton, S.A.; Schepens, B.; Bruynooghe, Y.; Beyaert, R.; Cornelis, S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: Roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2α. Biochem. J 2005, 385, 155–163. [Google Scholar]
- Schepens, B.; Tinton, S.A.; Bruynooghe, Y.; Parthoens, E.; Haegman, M.; Beyaert, R.; Cornelis, S. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J 2007, 26, 158–169. [Google Scholar]
- Damiano, F.; Rochira, A.; Tocci, R.; Alemanno, S.; Gnoni, A.; Siculella, L. hnRNP A1 mediates the activation of the IRES-dependent SREBP-1a mRNA translation in response to endoplasmic reticulum stress. Biochem. J 2013, 449, 543–553. [Google Scholar]
- Jo, O.D.; Martin, J.; Bernath, A.; Masri, J.; Lichtenstein, A.; Gera, J. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J. Biol. Chem 2008, 283, 23274–23287. [Google Scholar]
- Lewis, S.M.; Veyrier, A.; Hosszu Ungureanu, N.; Bonnal, S.; Vagner, S.; Holcik, M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol. Biol. Cell 2007, 18, 1302–1311. [Google Scholar]
- Cammas, A.; Pileur, F.; Bonnal, S.; Lewis, S.M.; Leveque, N.; Holcik, M.; Vagner, S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell 2007, 18, 5048–5059. [Google Scholar]
- Cloninger, C.; Bernath, A.; Bashir, T.; Holmes, B.; Artinian, N.; Ruegg, T.; Anderson, L.; Masri, J.; Lichtenstein, A.; Gera, J. Inhibition of SAPK2/p38 enhances sensitivity to mTORC1 inhibition by blocking IRES-mediated translation initiation in glioblastoma. Mol. Cancer Ther 2011, 10, 2244–2256. [Google Scholar]
- Henis-Korenblit, S.; Shani, G.; Sines, T.; Marash, L.; Shohat, G.; Kimchi, A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. USA 2002, 99, 5400–5405. [Google Scholar]
- Durie, D.; Lewis, S.M.; Liwak, U.; Kisilewicz, M.; Gorospe, M.; Holcik, M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene 2011, 30, 1460–1469. [Google Scholar]
- Lewis, S.M.; Cerquozzi, S.; Graber, T.E.; Ungureanu, N.H.; Andrews, M.; Holcik, M. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res 2008, 36, 168–178. [Google Scholar]
- Sammons, M.A.; Antons, A.K.; Bendjennat, M.; Udd, B.; Krahe, R.; Link, A.J. ZNF9 activation of IRES-mediated translation of the human ODC mRNA is decreased in myotonic dystrophy type 2. PLoS One 2010, 5, e9301. [Google Scholar]
- Faye, M.D.; Graber, T.E.; Liu, P.; Thakor, N.; Baird, S.D.; Durie, D.; Holcik, M. Nucleotide composition of cellular internal ribosome entry sites defines dependence on NF45 and predicts a posttranscriptional mitotic regulon. Mol. Cell Biol 2013, 33, 307–318. [Google Scholar]
- Graber, T.E.; Baird, S.D.; Kao, P.N.; Mathews, M.B.; Holcik, M. NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ 2010, 17, 719–729. [Google Scholar]
- Yeh, C.H.; Hung, L.Y.; Hsu, C.; Le, S.Y.; Lee, P.T.; Liao, W.L.; Lin, Y.T.; Chang, W.C.; Tseng, J.T. RNA-binding protein HuR interacts with thrombomodulin 5′ untranslated region and represses internal ribosome entry site-mediated translation under IL-1β treatment. Mol. Biol. Cell 2008, 19, 3812–3822. [Google Scholar]
- De Vries, S.; Naarmann-de Vries, I.S.; Urlaub, H.; Lue, H.; Bernhagen, J.; Ostareck, D.H.; Ostareck-Lederer, A. Identification of DEAD-box RNA helicase 6 (DDX6) as a cellular modulator of vascular endothelial growth factor expression under hypoxia. J. Biol. Chem 2013, 288, 5815–5827. [Google Scholar]
- Ungureanu, N.H.; Cloutier, M.; Lewis, S.M.; de Silva, N.; Blais, J.D.; Bell, J.C.; Holcik, M. Internal ribosome entry site-mediated translation of Apaf-1, but not XIAP, is regulated during UV-induced cell death. J. Biol. Chem 2006, 281, 15155–15163. [Google Scholar]
- Liwak, U.; Thakor, N.; Jordan, L.E.; Roy, R.; Lewis, S.M.; Pardo, O.E.; Seckl, M.; Holcik, M. Tumor suppressor PDCD4 represses internal ribosome entry site-mediated translation of antiapoptotic proteins and is regulated by S6 kinase 2. Mol. Cell Biol 2012, 32, 1818–1829. [Google Scholar]
- Rocchi, L.; Pacilli, A.; Sethi, R.; Penzo, M.; Schneider, R.J.; Trere, D.; Brigotti, M.; Montanaro, L. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res 2013, 41, 8308–8318. [Google Scholar]
- Gerbasi, V.R.; Link, A.J. The myotonic dystrophy type 2 protein ZNF9 is part of an ITAF complex that promotes cap-independent translation. Mol. Cell Proteomics 2007, 6, 1049–1058. [Google Scholar]
- Hertz, M.I.; Landry, D.M.; Willis, A.E.; Luo, G.; Thompson, S.R. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell Biol 2013, 33, 1016–1026. [Google Scholar]
- Thompson, S.R. Tricks an IRES uses to enslave ribosomes. Trends Microbiol 2012, 20, 558–566. [Google Scholar]
- Malygin, A.A.; Kossinova, O.A.; Shatsky, I.N.; Karpova, G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res 2013, 41, 8706–8714. [Google Scholar]
- Chappell, S.A.; Edelman, G.M.; Mauro, V.P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. USA 2000, 97, 1536–1541. [Google Scholar]
- Martinez-Azorin, F.; Remacha, M.; Martinez-Salas, E.; Ballesta, J.P. Internal translation initiation on the foot-and-mouth disease virus IRES is affected by ribosomal stalk conformation. FEBS Lett 2008, 582, 3029–3032. [Google Scholar]
- Yang, C.; Zhang, C.; Dittman, J.D.; Whitham, S.A. Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology 2009, 390, 163–173. [Google Scholar]
- Basu, A.; Das, P.; Chaudhuri, S.; Bevilacqua, E.; Andrews, J.; Barik, S.; Hatzoglou, M.; Komar, A.A.; Mazumder, B. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular Internal Ribosome Entry Sites. Mol. Cell Biol 2011, 31, 4482–4499. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).