Mechanisms by Which Licochalcone E Exhibits Potent Anti-Inflammatory Properties: Studies with Phorbol Ester-Treated Mouse Skin and Lipopolysaccharide-Stimulated Murine Macrophages
Abstract
:1. Introduction
2. Results and Discussion
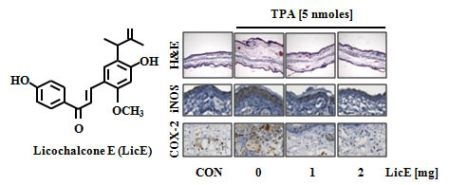
2.1. LicE Suppresses TPA-Induced Ear Edema in Mice
2.2. LicE Suppresses LPS-Induced mRNA Expression of Inflammatory Mediators and Enzymes, as well as iNOS and COX-2 Promoter Activities in RAW 264.7 Cells
2.3. LicE Suppresses LPS-Induced NF-κB Signaling in RAW 264.7 Cells
2.4. LicE Suppresses LPS-Induced AP-1 Signaling
2.5. Discussion
3. Experimental Section
3.1. Materials
3.2. In Vivo Inflammation Model
3.3. Immunohistochemistry (IHC)
3.4. Cell Culture and Assays of Cell Viability, NO, PGE2 and Cytokines
3.5. Western Blot Analysis
3.6. Real-Time RT-PCR
3.7. Luciferase Reporter Gene Assay
3.8. Electrophoretic Mobility Shift Assay (EMSA)
3.9. Statistical Analyses
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Henson, P.M. Dampening inflammation. Nat. Immunol 2005, 6, 1179–1181. [Google Scholar]
- Romero, R.; Espinoza, J.; Goncalves, L.F.; Kusanovic, J.P.; Friel, L.; Hassan, S. The role of inflammation and infection in preterm birth. Semin. Reprod. Med 2007, 25, 21–39. [Google Scholar]
- Ryan, G.B.; Majno, G. Acute inflammation. A review. Am. J. Pathol 1977, 86, 183–276. [Google Scholar]
- Prasad, S.; Sung, B.; Aggarwal, B.B. Age-associated chronic diseases require age-old medicine: Role of chronic inflammation. Prev. Med 2012, 54, S29–S37. [Google Scholar]
- Fujiwara, N.; Kobayashi, K. Macrophages in inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 281–286. [Google Scholar]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol 2003, 3, 23–35. [Google Scholar]
- Stuehr, D.J. Mammalian nitric oxide synthases. Biochim. Biophys. Acta 1999, 1411, 217–230. [Google Scholar]
- Korhonen, R.; Lahti, A.; Kankaanranta, H.; Moilanen, E. Nitric oxide production and signaling in inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 471–479. [Google Scholar]
- Liew, F.Y. Regulation of nitric oxide synthesis in infectious and autoimmune diseases. Immun. Lett 1994, 43, 95–98. [Google Scholar]
- Akarasereenont, P.; Bakhle, Y.S.; Thiemermann, C.; Vane, J.R. Cytokine-mediated induction of cyclo-oxygenase-2 by activation of tyrosine kinase in bovine endothelial cells stimulated by bacterial lipopolysaccharide. Br. J. Pharmacol 1995, 115, 401–408. [Google Scholar]
- Mitchell, J.A.; Belvisi, M.G.; Akarasereenont, P.; Robbins, R.A.; Kwon, O.J.; Croxtall, J.; Barnes, P.J.; Vane, J.R. Induction of cyclo-oxygenase-2 by cytokines in human pulmonary epithelial cells: regulation by dexamethasone. Br. J. Pharmacol 1994, 113, 1008–1014. [Google Scholar]
- Guha, M.; Mackman, N. LPS induction of gene expression in human monocytes. Cell Signal 2001, 13, 85–94. [Google Scholar]
- Li, Q.; Verma, I.M. NF-kappaB regulation in the immune system. Nat. Rev. Immunol 2002, 2, 725–734. [Google Scholar]
- Karin, M.; Ben-Neriah, Y. Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu. Rev. Immunol 2000, 18, 621–663. [Google Scholar]
- Karin, M. Inflammation-activated protein kinases as targets for drug development. Proc. Am. Thorac. Soc 2005, 2, 386–390, , discussion 394–395.. [Google Scholar]
- Matthews, C.P.; Colburn, N.H.; Young, M.R. AP-1 a target for cancer prevention. Curr. Cancer Drug Targets 2007, 7, 317–324. [Google Scholar]
- Xiao, W.; Hodge, D.R.; Wang, L.; Yang, X.; Zhang, X.; Farrar, W.L. NF-κB activates IL-6 expression through cooperation with c-Jun and IL6-AP1 site, but is independent of its IL6-NFkappaB regulatory site in autocrine human multiple myeloma cells. Cancer Biol. Ther 2004, 3, 1007–1017. [Google Scholar]
- Serkkola, E.; Hurme, M. Synergism between protein-kinase C and cAMP-dependent pathways in the expression of the interleukin-1 beta gene is mediated via the activator-protein-1 (AP-1) enhancer activity. Eur. J. Biochem 1993, 213, 243–249. [Google Scholar]
- Huang, K.C. Pharmacology of Chinese Herbs; CRC Press: Boca Raton, FL, USA, 1993. [Google Scholar]
- Kim, K.R.; Jeong, C.K.; Park, K.K.; Choi, J.H.; Park, J.H.; Lim, S.S.; Chung, W.Y. Anti-inflammatory effects of licorice and roasted licorice extracts on TPA-induced acute inflammation and collagen-induced arthritis in mice. J. Biomed. Biotechnol 2010, 2010, 709378. [Google Scholar]
- Chu, X.; Ci, X.; Wei, M.; Yang, X.; Cao, Q.; Guan, M.; Li, H.; Deng, Y.; Feng, H.; Deng, X. Licochalcone a inhibits lipopolysaccharide-induced inflammatory response in vitro and in vivo. J. Agric. Food Chem 2012, 60, 3947–3954. [Google Scholar]
- Kwon, H.S.; Park, J.H.; Kim, D.H.; Kim, Y.H.; Shin, H.K.; Kim, J.K. Licochalcone A isolated from licorice suppresses lipopolysaccharide-stimulated inflammatory reactions in RAW264.7 cells and endotoxin shock in mice. J. Mol. Med 2008, 86, 1287–1295. [Google Scholar]
- Yoon, G.; Jung, Y.D.; Cheon, S.H. Cytotoxic allyl retrochalcone from the roots of Glycyrrhiza inflata. Chem. Pharm. Bull 2005, 53, 694–695. [Google Scholar]
- Cho, Y.C.; Lee, S.H.; Yoon, G.; Kim, H.S.; Na, J.Y.; Choi, H.J.; Cho, C.W.; Cheon, S.H.; Kang, B.Y. Licochalcone E reduces chronic allergic contact dermatitis and inhibits IL-12p40 production through down-regulation of NF-kappa B. Int. Immunopharmacol 2010, 10, 1119–1126. [Google Scholar]
- Kwon, S.J.; Park, S.Y.; Kwon, G.T.; Lee, K.W.; Kang, Y.H.; Choi, M.S.; Yun, J.W.; Jeon, J.H.; Jun, J.G.; Park, J.H. Licochalcone E present in licorice suppresses lung metastasis in the 4T1 mammary orthotopic cancer model. Cancer Prev. Res. 2013. [Google Scholar] [CrossRef]
- Sommer, P.; Ray, D.W. Novel therapeutic agents targeting the glucocorticoid receptor for inflammation and cancer. Curr. Opin. Investig. Drugs 2008, 9, 1070–1077. [Google Scholar]
- Lukens, J.R.; Gross, J.M.; Kanneganti, T.D. IL-1 family cytokines trigger sterile inflammatory disease. Front. Immunol 2012, 3, 315. [Google Scholar]
- Gabay, C. Interleukin-6 and chronic inflammation. Arthritis. Res. Ther 2006, 8, S3. [Google Scholar]
- Locksley, R.M.; Killeen, N.; Lenardo, M.J. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell 2001, 104, 487–501. [Google Scholar]
- Park, G.M.; Jun, J.G.; Kim, J.K. Anti-inflammatory effect of licochalcone E, a constituent of licorice, on lipopolysaccharide-induced inflammatory responses in murine macrophages. J. Life Sci 2011, 21, 656–663. [Google Scholar]
- Ghosh, S.; Karin, M. Missing pieces in the NF-kappaB puzzle. Cell 2002, 109, S81–S96. [Google Scholar]
- Delhase, M.; Hayakawa, M.; Chen, Y.; Karin, M. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science 1999, 284, 309–313. [Google Scholar]
- Janssen-Heininger, Y.M.; Poynter, M.E.; Baeuerle, P.A. Recent advances towards understanding redox mechanisms in the activation of nuclear factor kappaB. Free Radic. Biol. Med 2000, 28, 1317–1327. [Google Scholar]
- Schulze-Osthoff, K.; Ferrari, D.; Riehemann, K.; Wesselborg, S. Regulation of NF-kappa B activation by MAP kinase cascades. Immunobiology 1997, 198, 35–49. [Google Scholar]
- Kane, L.P.; Shapiro, V.S.; Stokoe, D.; Weiss, A. Induction of NF-kappaB by the Akt/PKB kinase. Curr. Biol 1999, 9, 601–604. [Google Scholar]
- Chun, K.S.; Keum, Y.S.; Han, S.S.; Song, Y.S.; Kim, S.H.; Surh, Y.J. Curcumin inhibits phorbol ester-induced expression of cyclooxygenase-2 in mouse skin through suppression of extracellular signal-regulated kinase activity and NF-kappaB activation. Carcinogenesis 2003, 24, 1515–1524. [Google Scholar]
- Jeon, J.H.; Kim, M.R.; Kwon, E.M.; Lee, N.R.; Jun, J.G. Highly efficient synthesis of licochalcone E through water-accelerated [3,3]-sigmatropic rearrangement of allyl aryl ether. Bull. Korean Chem. Soc 2011, 32, 1059–1062. [Google Scholar]
- Park, H.; Kim, M.; Kwon, G.T.; Lim do, Y.; Yu, R.; Sung, M.K.; Lee, K.W.; Daily, J.W., 3rd; Park, J.H. A high-fat diet increases angiogenesis, solid tumor growth, and lung metastasis of CT26 colon cancer cells in obesity-resistant BALB/c mice. Mol. Carcinog 2012, 51, 869–880. [Google Scholar]
- Cho, H.J.; Seon, M.R.; Lee, Y.M.; Kim, J.; Kim, J.K.; Kim, S.G.; Park, J.H. 3,3′-Diindolylmethane suppresses the inflammatory response to lipopolysaccharide in murine macrophages. J. Nutr 2008, 138, 17–23. [Google Scholar]
- Shin, N.R.; Lee, D.Y.; Shin, S.J.; Kim, K.S.; Yoo, H.S. Regulation of proinflammatory mediator production in RAW264.7 macrophage by Vibrio vulnificus luxS and smcR. FEMS Immunol. Med. Microbiol 2004, 41, 169–176. [Google Scholar]
- Zhao, L.; Zhang, S.L.; Tao, J.Y.; Pang, R.; Jin, F.; Guo, Y.J.; Dong, J.H.; Ye, P.; Zhao, H.Y.; Zheng, G.H. Preliminary exploration on anti-inflammatory mechanism of Corilagin (beta-1-O-galloyl-3,6-(R)-hexahydroxydiphenoyl-D-glucose) in vitro. Int. Immunopharmacol 2008, 8, 1059–1064. [Google Scholar]
- Cho, I.J.; Lee, A.K.; Lee, S.J.; Lee, M.G.; Kim, S.G. Repression by oxidative stress of iNOS and cytokine gene induction in macrophages results from AP-1 and NF-kappaB inhibition mediated by B cell translocation gene-1 activation. Free Radic. Biol. Med 2005, 39, 1523–1536. [Google Scholar]
- Ki, S.H.; Choi, M.J.; Lee, C.H.; Kim, S.G. Galpha12 specifically regulates COX-2 induction by sphingosine 1-phosphate. Role for JNK-dependent ubiquitination and degradation of IkappaBalpha. J. Biol. Chem 2007, 282, 1938–1947. [Google Scholar]
- De Wet, J.R.; Wood, K.V.; DeLuca, M.; Helinski, D.R.; Subramani, S. Firefly luciferase gene: Structure and expression in mammalian cells. Mol. Cell Biol 1987, 7, 725–737. [Google Scholar]










© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lee, H.N.; Cho, H.J.; Lim, D.Y.; Kang, Y.-H.; Lee, K.W.; Park, J.H.Y. Mechanisms by Which Licochalcone E Exhibits Potent Anti-Inflammatory Properties: Studies with Phorbol Ester-Treated Mouse Skin and Lipopolysaccharide-Stimulated Murine Macrophages. Int. J. Mol. Sci. 2013, 14, 10926-10943. https://doi.org/10.3390/ijms140610926
Lee HN, Cho HJ, Lim DY, Kang Y-H, Lee KW, Park JHY. Mechanisms by Which Licochalcone E Exhibits Potent Anti-Inflammatory Properties: Studies with Phorbol Ester-Treated Mouse Skin and Lipopolysaccharide-Stimulated Murine Macrophages. International Journal of Molecular Sciences. 2013; 14(6):10926-10943. https://doi.org/10.3390/ijms140610926
Chicago/Turabian StyleLee, Han Na, Han Jin Cho, Do Young Lim, Young-Hee Kang, Ki Won Lee, and Jung Han Yoon Park. 2013. "Mechanisms by Which Licochalcone E Exhibits Potent Anti-Inflammatory Properties: Studies with Phorbol Ester-Treated Mouse Skin and Lipopolysaccharide-Stimulated Murine Macrophages" International Journal of Molecular Sciences 14, no. 6: 10926-10943. https://doi.org/10.3390/ijms140610926
APA StyleLee, H. N., Cho, H. J., Lim, D. Y., Kang, Y. -H., Lee, K. W., & Park, J. H. Y. (2013). Mechanisms by Which Licochalcone E Exhibits Potent Anti-Inflammatory Properties: Studies with Phorbol Ester-Treated Mouse Skin and Lipopolysaccharide-Stimulated Murine Macrophages. International Journal of Molecular Sciences, 14(6), 10926-10943. https://doi.org/10.3390/ijms140610926