Regulatory Non-Coding RNAs in Pluripotent Stem Cells
Abstract
:1. Introduction
2. MicroRNAs in Embryonic Stem Cells
2.1. miRNAs Regulate ESC Cell Cycle
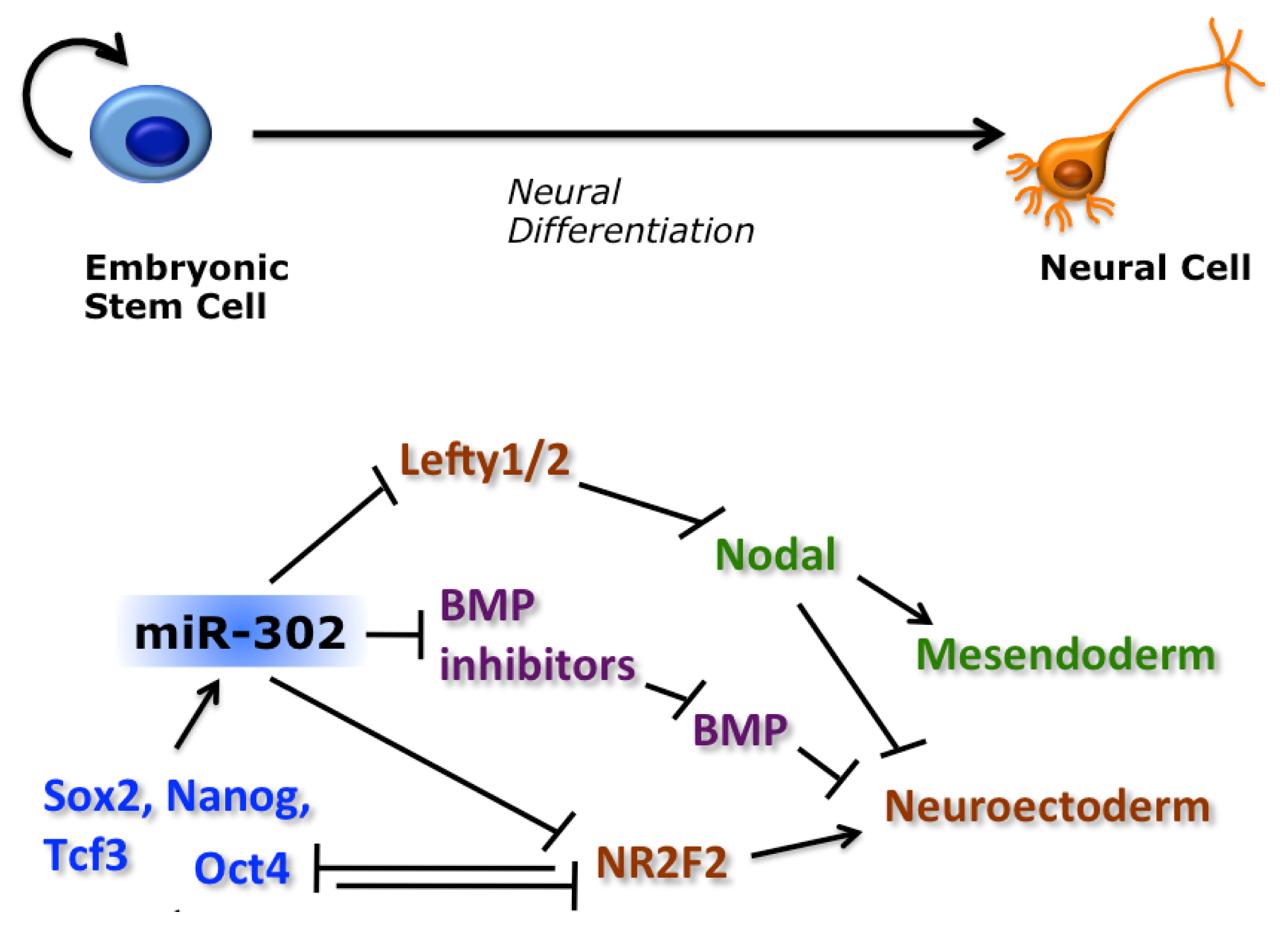
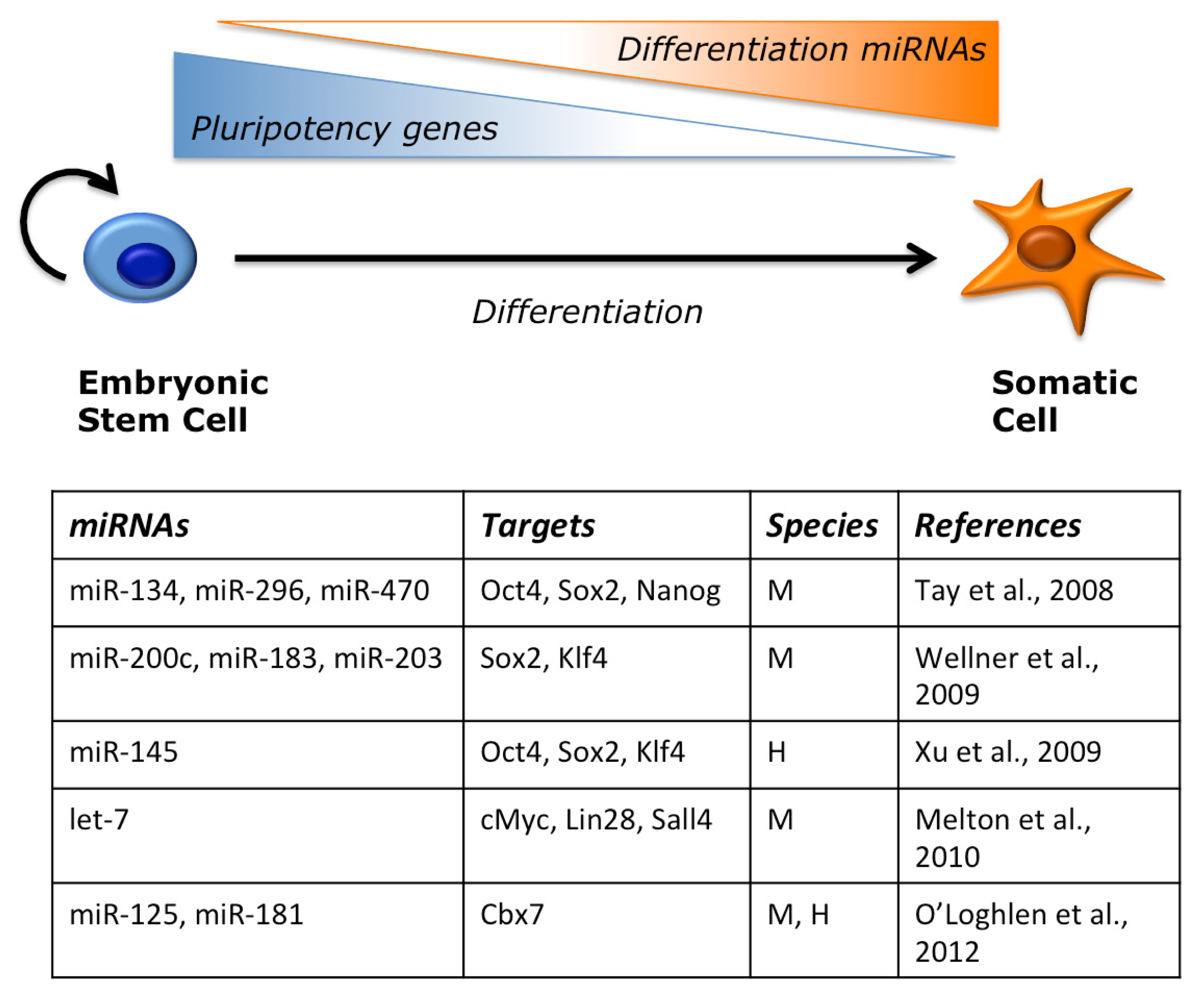
2.2. miRNAs Regulate ESC Pluripotency and Differentiation
3. MicroRNAs and Reprogramming to iPSCs
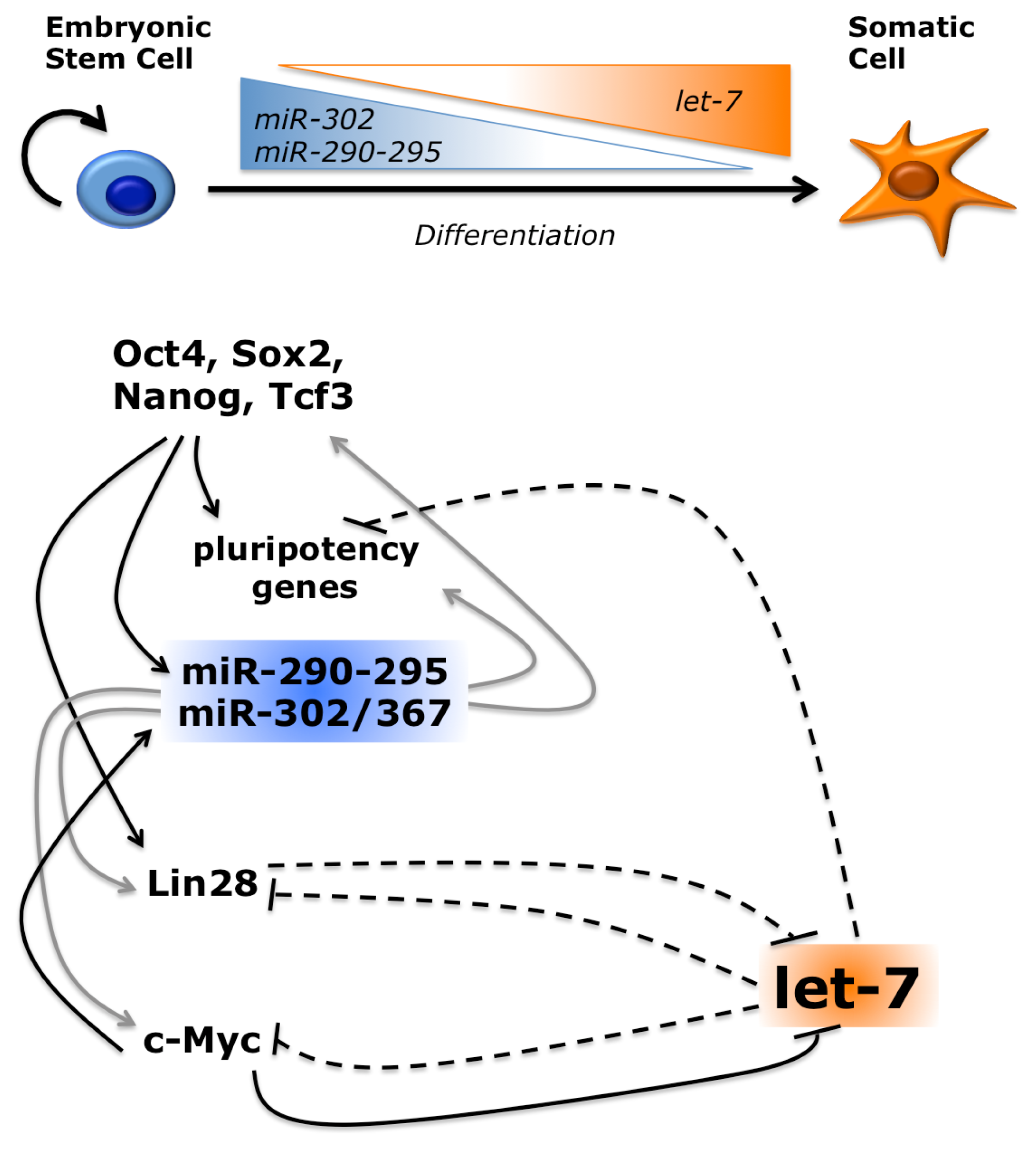
3.1. Several miRNAs Promote or Inhibit Reprogramming
3.2. miRNAs Reprogram Somatic Cells in the Absence of Protein-Coding Reprogramming Factors
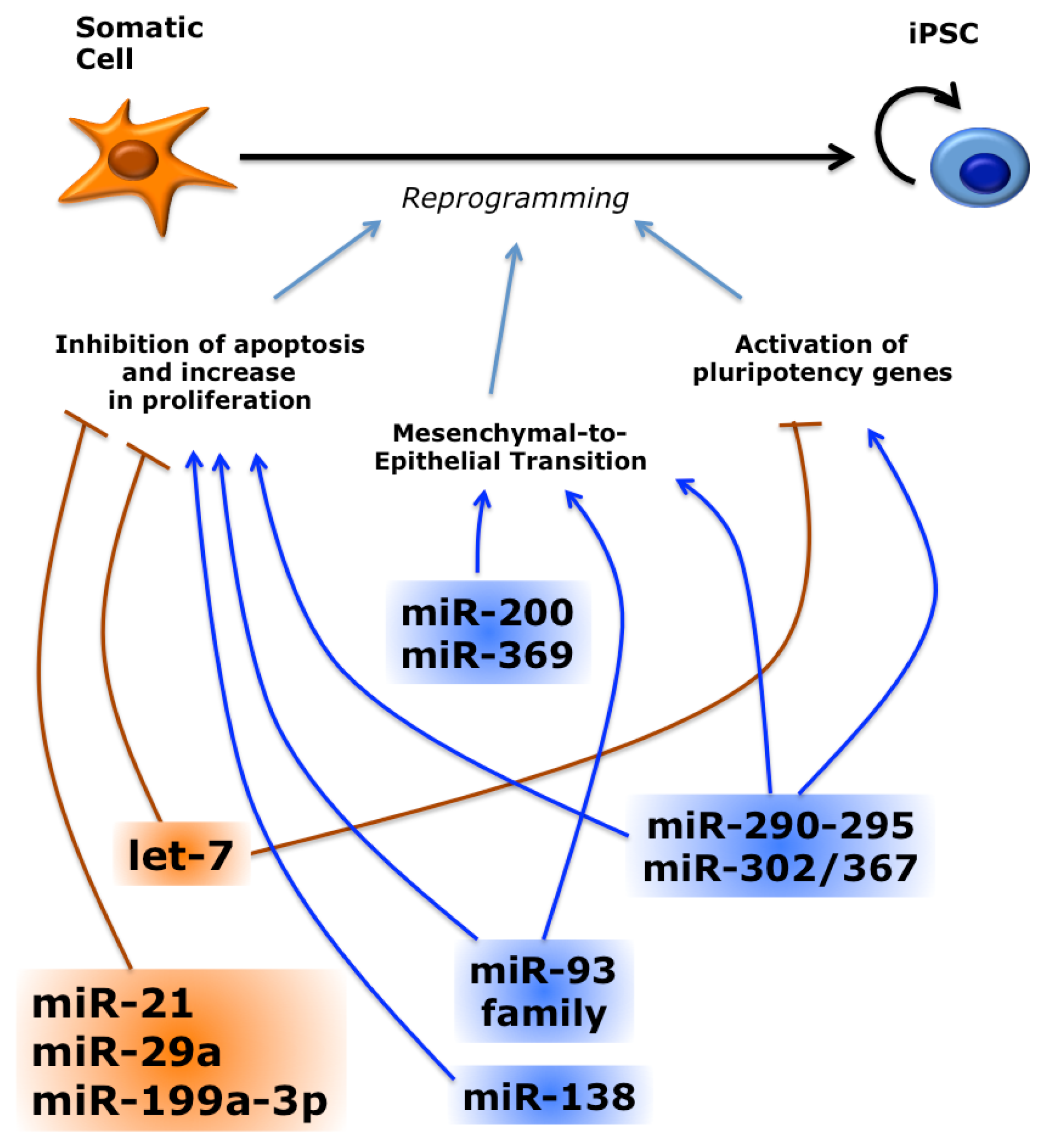
3.3. miRNAs Regulate Reprogramming by Multiple Pathways
4. Long Non-Coding RNAs in Embryonic Stem Cells
lncRNAs Maintain Pluripotency in ESC
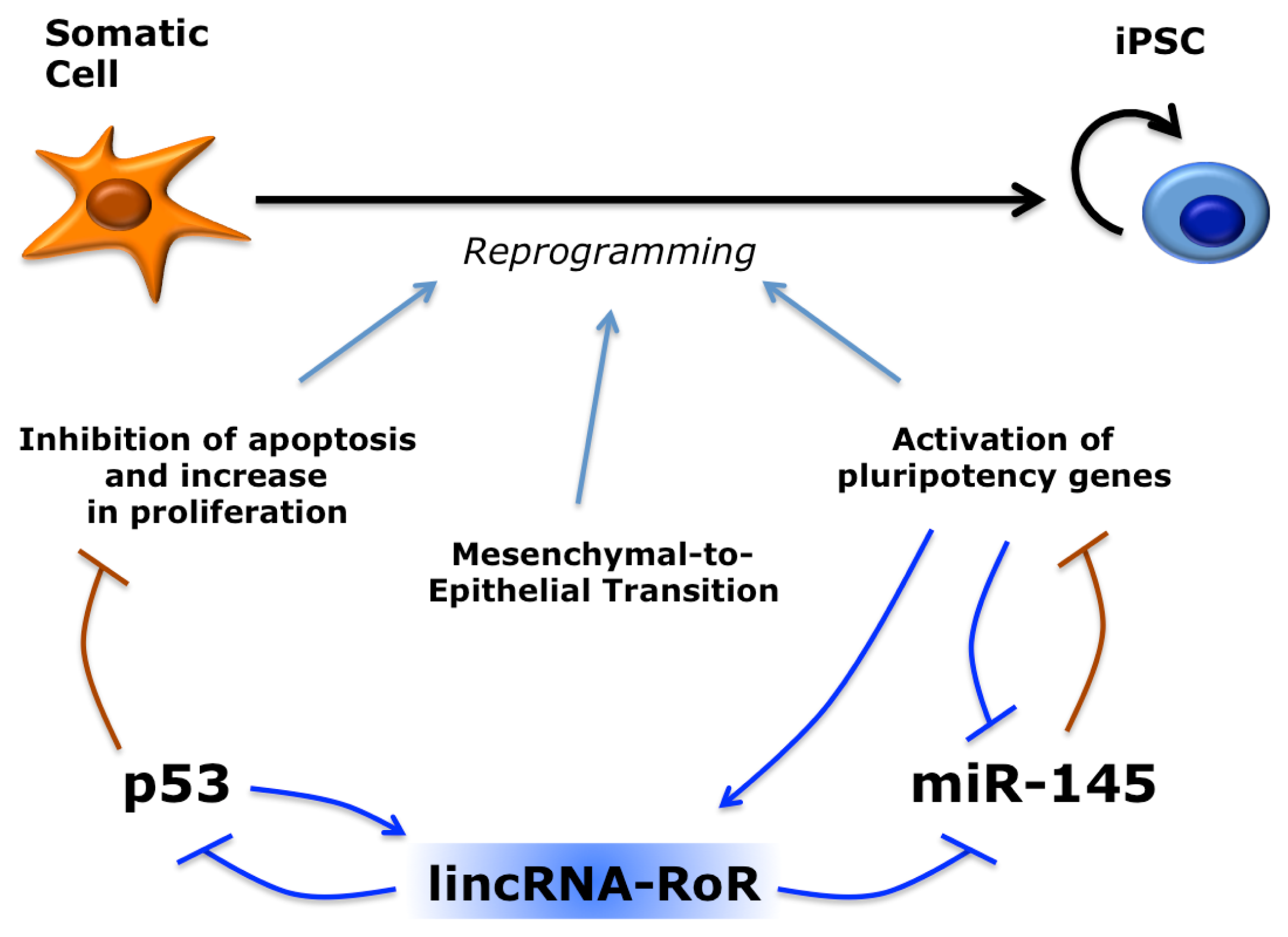
5. Long Non-Coding RNAs in Induced Pluripotent Stem Cells
6. Conclusions and Future Perspectives
Acknowledgments
Conflict of Interest
References
- Warmflash, A.; Arduini, B.L.; Brivanlou, A.H. The molecular circuitry underlying pluripotency in embryonic stem cells. Wiley Interdiscip. Rev. Syst. Biol. Med 2012, 4, 443–456. [Google Scholar]
- James, D.; Levine, A.J.; Besser, D.; Hemmati-Brivanlou, A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 2005, 132, 1273–1282. [Google Scholar]
- Vallier, L.; Alexander, M.; Pedersen, R.A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci 2005, 118, 4495–4509. [Google Scholar]
- Ying, Q.L.; Nichols, J.; Chambers, I.; Smith, A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003, 115, 281–292. [Google Scholar]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar]
- Kim, J.; Chu, J.; Shen, X.; Wang, J.; Orkin, S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 2008, 132, 1049–1061. [Google Scholar]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B.; Wong, E.; Orlov, Y.L.; Zhang, W.; Jiang, J.; et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008, 133, 1106–1117. [Google Scholar]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar]
- Spivakov, M.; Fisher, A.G. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet 2007, 8, 263–271. [Google Scholar]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar]
- Guo, G.; Yang, J.; Nichols, J.; Hall, J.S.; Eyres, I.; Mansfield, W.; Smith, A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 2009, 136, 1063–1069. [Google Scholar]
- Silva, J.; Nichols, J.; Theunissen, T.W.; Guo, G.; van Oosten, A.L.; Barrandon, O.; Wray, J.; Yamanaka, S.; Chambers, I.; Smith, A. Nanog is the gateway to the pluripotent ground state. Cell 2009, 138, 722–737. [Google Scholar]
- Pera, M.F.; Tam, P.P.L. Extrinsic regulation of pluripotent stem cells. Nature 2010, 465, 713–720. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar]
- Stadtfeld, M.; Hochedlinger, K. Induced pluripotency: History, mechanisms, and applications. Genes Dev 2010, 24, 2239–2263. [Google Scholar]
- Papp, B.; Plath, K. Reprogramming to pluripotency: Stepwise resetting of the epigenetic landscape. Cell Res 2011, 21, 486–501. [Google Scholar]
- Cohen, D.E.; Melton, D. Turning straw into gold: Directing cell fate for regenerative medicine. Nat. Rev. Genet 2011, 12, 243–252. [Google Scholar]
- Mattick, J.S. The central role of RNA in human development and cognition. FEBS Lett 2011, 585, 1600–1616. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol 2007, 23, 175–205. [Google Scholar]
- Fatica, A.; Rosa, A.; Ballarino, M.; de Marchis, M.L.; Rasmussen, K.D.; Bozzoni, I. Role of microRNAs in myeloid differentiation. Biochem. Soc. Trans 2008, 36, 1201–1205. [Google Scholar]
- Rosa, A.; Brivanlou, A.H. microRNAs in early vertebrate development. Cell Cycle 2009, 8, 3531–3520. [Google Scholar]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol 2012, 13, 239–250. [Google Scholar]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med 2012, 4, 143–159. [Google Scholar]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.E.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar]
- Hammond, S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett 2005, 579, 5822–5829. [Google Scholar]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar]
- Rajewsky, N. microRNA target predictions in animals. Nat. Genet 2006, 38, S8–S13. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci 2007, 120, 2498–2506. [Google Scholar]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Soldà, G.; Simons, C.; et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008, 18, 1433–1445. [Google Scholar]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C.; et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol 2010, 28, 503–510. [Google Scholar]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar]
- Nakaya, H.I.; Amaral, P.P.; Louro, R.; Lopes, A.; Fachel, A.A.; Moreira, Y.B.; El-Jundi, T.A.; da Silva, A.M.; Reis, E.M.; Verjovski-Almeida, S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol 2007, 8, R43. [Google Scholar]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar]
- Ravasi, T.; Suzuki, H.; Pang, K.C.; Katayama, S.; Furuno, M.; Okunishi, R.; Fukuda, S.; Ru, K.; Frith, M.C.; Gongora, M.M.; et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 2006, 16, 11–19. [Google Scholar]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 2007, 17, 556–565. [Google Scholar]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar]
- Mattick, J.S. A new paradigm for developmental biology. J. Exp. Biol 2007, 210, 1526–1547. [Google Scholar]
- Carninci, P.; Hayashizaki, Y. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev 2007, 17, 139–144. [Google Scholar]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar]
- Beltran, M.; Puig, I.; Peña, C.; García, J.M.; Alvarez, A.B.; Peña, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 2008, 22, 756–769. [Google Scholar]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific microRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar]
- Laurent, L.C.; Chen, J.; Ulitsky, I.; Mueller, F.-J.; Lu, C.; Shamir, R.; Fan, J.-B.; Loring, J.F. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 2008, 26, 1506–1516. [Google Scholar]
- Bar, M.; Wyman, S.K.; Fritz, B.R.; Qi, J.; Garg, K.S.; Parkin, R.K.; Kroh, E.M.; Bendoraite, A.; Mitchell, P.S.; Nelson, A.M.; et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 2008, 26, 2496–2505. [Google Scholar]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 2008, 18, 610–621. [Google Scholar]
- Ciaudo, C.; Servant, N.; Cognat, V.; Sarazin, A.; Kieffer, E.; Viville, S.; Colot, V.; Barillot, E.; Heard, E.; Voinnet, O. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet 2009, 5, e1000620. [Google Scholar]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar]
- Calabrese, J.M.; Seila, A.C.; Yeo, G.W.; Sharp, P.A. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 18097–18102. [Google Scholar]
- Lipchina, I.; Elkabetz, Y.; Hafner, M.; Sheridan, R.; Mihailovic, A.; Tuschl, T.; Sander, C.; Studer, L.; Betel, D. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev 2011, 25, 2173–2186. [Google Scholar]
- Barroso-delJesus, A.; Romero-López, C.; Lucena-Aguilar, G.; Melen, G.J.; Sanchez, L.; Ligero, G.; Berzal-Herranz, A.; Menendez, P. Embryonic stem cell-specific miR302-367 cluster: Human gene structure and functional characterization of its core promoter. Mol. Cell Biol 2008, 28, 6609–6619. [Google Scholar]
- Card, D.A.G.; Hebbar, P.B.; Li, L.; Trotter, K.W.; Komatsu, Y.; Mishina, Y.; Archer, T.K. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol 2008, 28, 6426–6438. [Google Scholar]
- Jouneau, A.; Ciaudo, C.; Sismeiro, O.; Brochard, V.; Jouneau, L.; Vandormael-Pournin, S.; Coppée, J.-Y.; Zhou, Q.; Heard, E.; Antoniewski, C.; et al. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA 2012, 18, 253–264. [Google Scholar]
- Stadler, B.; Ivanovska, I.; Mehta, K.; Song, S.; Nelson, A.; Tan, Y.; Mathieu, J.; Darby, C.; Blau, C.A.; Ware, C.; et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev 2010, 19, 935–950. [Google Scholar]
- Hanna, J.; Cheng, A.W.; Saha, K.; Kim, J.; Lengner, C.J.; Soldner, F.; Cassady, J.P.; Muffat, J.; Carey, B.W.; Jaenisch, R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA 2010, 107, 9222–9227. [Google Scholar]
- Kim, H.; Lee, G.; Ganat, Y.; Papapetrou, E.P.; Lipchina, I.; Socci, N.D.; Sadelain, M.; Studer, L. miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell 2011, 8, 695–706. [Google Scholar]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet 2003, 35, 215–217. [Google Scholar]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005, 19, 489–501. [Google Scholar]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar]
- Wang, Y.; Medvid, R.; Melton, C.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet 2007, 39, 380–385. [Google Scholar]
- Sinkkonen, L.; Hugenschmidt, T.; Berninger, P.; Gaidatzis, D.; Mohn, F.; Artus-Revel, C.G.; Zavolan, M.; Svoboda, P.; Filipowicz, W. microRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol 2008, 15, 259–267. [Google Scholar]
- Wang, Y.; Baskerville, S.; Shenoy, A.; Babiarz, J.E.; Baehner, L.; Blelloch, R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet 2008, 40, 1478–1483. [Google Scholar]
- Wang, Y.; Blelloch, R. Cell cycle regulation by microRNAs in embryonic stem cells. Cancer Res 2009, 69, 4093–4096. [Google Scholar]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Muñoz, P.; Gonzalez, S.; Schoeftner, S.; Murchison, E.; Andl, T.; Chen, T.; Klatt, P.; et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol 2008, 15, 268–279. [Google Scholar]
- Kim, B.-M.; Choi, M.Y. Non-canonical microRNAs miR-320 and miR-702 promote proliferation in Dgcr8-deficient embryonic stem cells. Biochem. Biophys. Res. Commun 2012, 426, 183–189. [Google Scholar]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008, 22, 2773–2785. [Google Scholar]
- White, J.; Dalton, S. Cell cycle control of embryonic stem cells. Stem Cell Rev 2005, 1, 131–138. [Google Scholar]
- Qi, J.; Yu, J.-Y.; Shcherbata, H.R.; Mathieu, J.; Wang, A.J.; Seal, S.; Zhou, W.; Stadler, B.M.; Bourgin, D.; Wang, L.; et al. microRNAs regulate human embryonic stem cell division. Cell Cycle 2009, 8, 3729–3741. [Google Scholar]
- Dolezalova, D.; Mraz, M.; Barta, T.; Plevova, K.; Vinarsky, V.; Holubcova, Z.; Jaros, J.; Dvorak, P.; Pospisilova, S.; Hampl, A. microRNAs regulate p21(Waf1/Cip1) protein expression and the DNA damage response in human embryonic stem cells. Stem Cells 2012, 30, 1362–1372. [Google Scholar]
- Sengupta, S.; Nie, J.; Wagner, R.J.; Yang, C.; Stewart, R.; Thomson, J.A. microRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells 2009, 27, 1524–1528. [Google Scholar]
- Lee, N.S.; Kim, J.S.; Cho, W.J.; Lee, M.R.; Steiner, R.; Gompers, A.; Ling, D.; Zhang, J.; Strom, P.; Behlke, M.; et al. miR-302b maintains “stemness” of human embryonal carcinoma cells by post-transcriptional regulation of Cyclin D2 expression. Biochem. Biophys. Res. Commun 2008, 377, 434–440. [Google Scholar]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.-C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar]
- Lipchina, I.; Studer, L.; Betel, D. The expanding role of miR-302-367 in pluripotency and reprogramming. Cell Cycle 2012, 11, 1517–1523. [Google Scholar]
- Smith, J.R.; Vallier, L.; Lupo, G.; Alexander, M.; Harris, W.A.; Pedersen, R.A. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev. Biol 2008, 313, 107–117. [Google Scholar]
- Vallier, L.; Reynolds, D.; Pedersen, R.A. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev. Biol 2004, 275, 403–421. [Google Scholar]
- Rosa, A.; Spagnoli, F.M.; Brivanlou, A.H. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 2009, 16, 517–527. [Google Scholar]
- Choi, W.-Y.; Giraldez, A.J.; Schier, A.F. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 2007, 318, 271–274. [Google Scholar]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol 2009, 27, 275–280. [Google Scholar]
- Ozair, M.Z.; Noggle, S.; Warmflash, A.; Krzyspiak, J.E.; Brivanlou, A.H. SMAD7 Directly converts human embryonic stem cells to telencephalic fate by a default mechanism. Stem Cells 2013, 31, 35–47. [Google Scholar]
- Kang, H.; Louie, J.; Weisman, A.; Sheu-Gruttadauria, J.; Davis-Dusenbery, B.N.; Lagna, G.; Hata, A. Inhibition of microRNA-302 (miR-302) by bone morphogenetic protein 4 (BMP4) facilitates the BMP signaling pathway. J. Biol. Chem 2012, 287, 38656–38664. [Google Scholar]
- Parisi, S.; Battista, M.; Musto, A.; Navarra, A.; Tarantino, C.; Russo, T. A regulatory loop involving Dies1 and miR-125a controls BMP4 signaling in mouse embryonic stem cells. FASEB J 2012, 26, 3957–3968. [Google Scholar]
- Zhou, A.-D.; Diao, L.-T.; Xu, H.; Xiao, Z.-D.; Li, J.-H.; Zhou, H.; Qu, L.-H. β-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/β-catenin-signaling pathway. Oncogene 2012, 31, 2968–2978. [Google Scholar]
- Rosa, A.; Brivanlou, A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J 2010, 30, 237–248. [Google Scholar]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. microRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol 2009, 11, 1487–1495. [Google Scholar]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing “stemness”. Nature 2008, 452, 225–229. [Google Scholar]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.; Kosik, K. microRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar]
- Morey, L.; Pascual, G.; Cozzuto, L.; Roma, G.; Wutz, A.; Benitah, S.A.; Di Croce, L. Nonoverlapping functions of the polycomb group cbx family of proteins in embryonic stem cells. Cell Stem Cell 2012, 10, 47–62. [Google Scholar]
- O’Loghlen, A.; Muñoz-Cabello, A.M.; Gaspar-Maia, A.; Wu, H.-A.; Banito, A.; Kunowska, N.; Racek, T.; Pemberton, H.N.; Beolchi, P.; Lavial, F.; et al. MicroRNA regulation of Cbx7 mediates a switch of polycomb orthologs during ESC differentiation. Cell Stem Cell 2012, 10, 33–46. [Google Scholar]
- Ambros, V. The evolution of our thinking about microRNAs. Nat. Med 2008, 14, 1036–1040. [Google Scholar]
- Büssing, I.; Slack, F.J.; Großhans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med 2008, 14, 400–409. [Google Scholar]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006, 20, 2202–2207. [Google Scholar]
- Viswanathan, S.R.; Daley, G.Q.; Gregory, R.I. Selective blockade of microRNA processing by Lin28. Science 2008, 320, 97–100. [Google Scholar]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A microRNA regulator with a macro role. Cell 2010, 140, 445–449. [Google Scholar]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar]
- Heo, I.; Joo, C.; Kim, Y.-K.; Ha, M.; Yoon, M.-J.; Cho, J.; Yeom, K.-H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar]
- Hagan, J.P.; Piskounova, E.; Gregory, R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol 2009, 16, 1021–1025. [Google Scholar]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol 2008, 10, 987–993. [Google Scholar]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar]
- Cartwright, P.; McLean, C.; Sheppard, A.; Rivett, D.; Jones, K.; Dalton, S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 2005, 132, 885–896. [Google Scholar]
- Lin, C.-H.; Jackson, A.L.; Guo, J.; Linsley, P.S.; Eisenman, R.N. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J 2009, 28, 3157–3170. [Google Scholar]
- Chang, T.-C.; Yu, D.; Lee, Y.-S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet 2008, 40, 43–50. [Google Scholar]
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar]
- Wilson, K.D.; Venkatasubrahmanyam, S.; Jia, F.; Sun, N.; Butte, A.J.; Wu, J.C. microRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev 2009, 18, 749–757. [Google Scholar]
- Neveu, P.; Kye, M.J.; Qi, S.; Buchholz, D.E.; Clegg, D.O.; Sahin, M.; Park, I.-H.; Kim, K.-S.; Daley, G.Q.; Kornblum, H.I.; et al. MicroRNA profiling reveals two distinct p53-related human pluripotent stem cell states. Cell Stem Cell 2010, 7, 671–681. [Google Scholar]
- Stadtfeld, M.; Apostolou, E.; Akutsu, H.; Fukuda, A.; Follett, P.; Natesan, S.; Kono, T.; Shioda, T.; Hochedlinger, K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 2010, 465, 175–181. [Google Scholar]
- Peters, J.; Robson, J.E. Imprinted noncoding RNAs. Mamm. Genome 2008, 19, 493–502. [Google Scholar]
- Li, Z.; Yang, C.-S.; Nakashima, K.; Rana, T.M. Small RNA-mediated regulation of iPS cell generation. EMBO J 2011, 30, 823–834. [Google Scholar]
- Judson, R.; Babiarz, J.; Venere, M.; Blelloch, R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol 2009, 27, 459–461. [Google Scholar]
- Liao, B.; Bao, X.; Liu, L.; Feng, S.; Zovoilis, A.; Liu, W.; Xue, Y.; Cai, J.; Guo, X.; Qin, B.; et al. MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem 2011, 286, 17359–17364. [Google Scholar]
- Ye, D.; Wang, G.; Liu, Y.; Huang, W.; Wu, M.; Zhu, S.; Jia, W.; Deng, A.-M.; Liu, H.; Kang, J. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells 2012, 30, 1645–1654. [Google Scholar]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol 2011, 29, 443–448. [Google Scholar]
- Hanna, J.; Saha, K.; Pando, B.; van Zon, J.; Lengner, C.J.; Creyghton, M.P.; van Oudenaarden, A.; Jaenisch, R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 2009, 462, 595–601. [Google Scholar]
- Yang, C.-S.; Li, Z.; Rana, T.M. microRNAs modulate iPS cell generation. RNA 2011, 17, 1451–1460. [Google Scholar]
- Wang, J.; He, Q.; Han, C.; Gu, H.; Jin, L.; Li, Q.; Mei, Y.; Wu, M. p53-facilitated miR-199a-3p regulates somatic cell reprogramming. Stem Cells 2012, 30, 1405–1413. [Google Scholar]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A.; Morrisey, E.E. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F.; et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar]
- Lin, S.-L.; Chang, D.C.; Chang-Lin, S.; Lin, C.-H.; Wu, D.T.S.; Chen, D.T.; Ying, S.-Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14, 2115–2124. [Google Scholar]
- Lin, S.-L.; Chang, D.C.; Lin, C.-H.; Ying, S.-Y.; Leu, D.; Wu, D.T.S. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res 2011, 39, 1054–1065. [Google Scholar]
- Menendez, S.; Camus, S.; Izpisúa Belmonte, J.C. p53: Guardian of reprogramming. Cell Cycle 2010, 9, 3887–3891. [Google Scholar]
- Jain, A.K.; Allton, K.; Iacovino, M.; Mahen, E.; Milczarek, R.J.; Zwaka, T.P.; Kyba, M.; Barton, M.C. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol 2012, 10, e1001268. [Google Scholar]
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63. [Google Scholar]
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.-K.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010, 7, 64–77. [Google Scholar]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol 2012, 13, 616–630. [Google Scholar]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol 2008, 10, 593–601. [Google Scholar]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 2008, 9, 582–589. [Google Scholar]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 2008, 68, 7846–7854. [Google Scholar]
- Barroso-delJesus, A.; Lucena-Aguilar, G.; Sanchez, L.; Ligero, G.; Gutierrez-Aranda, I.; Menendez, P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J 2011, 25, 1497–1508. [Google Scholar]
- Lee, M.R.; Prasain, N.; Chae, H.-D.; Kim, Y.-J.; Mantel, C.; Yoder, M.C.; Broxmeyer, H.E. Epigenetic regulation of nanog by MiR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem Cells 2012, 31, 666–681. [Google Scholar]
- Hu, S.; Wilson, K.D.; Ghosh, Z.; Han, L.; Wang, Y.; Lan, F.; Ransohoff, K.J.; Burridge, P.; Wu, J.C. microRNA-302 increases reprogramming efficiency via repression of NR2F2. Stem Cells 2013, 31, 259–268. [Google Scholar]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar]
- Sheik Mohamed, J.; Gaughwin, P.M.; Lim, B.; Robson, P.; Lipovich, L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 2010, 16, 324–337. [Google Scholar]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [Green Version]
- Ng, S.-Y.; Johnson, R.; Stanton, L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 2011, 31, 522–533. [Google Scholar]
- Loewer, S.; Cabili, M.N.; Guttman, M.; Loh, Y.-H.; Thomas, K.; Park, I.H.; Garber, M.; Curran, M.; Onder, T.; Agarwal, S.; et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet 2010, 42, 1113–1117. [Google Scholar] [Green Version]
- Hawkins, P.G.; Morris, K.V. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription 2010, 1, 165–175. [Google Scholar]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar]
- Zhang, A.; Zhou, N.; Huang, J.; Liu, Q.; Fukuda, K.; Ma, D.; Lu, Z.; Bai, C.; Watabe, K.; Mo, Y.-Y. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res 2012, 23, 340–350. [Google Scholar]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. microRNA-mediated conversion of human fibroblasts to neurons. Nature 2011, 476, 228–231. [Google Scholar]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar]


© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rosa, A.; Brivanlou, A.H. Regulatory Non-Coding RNAs in Pluripotent Stem Cells. Int. J. Mol. Sci. 2013, 14, 14346-14373. https://doi.org/10.3390/ijms140714346
Rosa A, Brivanlou AH. Regulatory Non-Coding RNAs in Pluripotent Stem Cells. International Journal of Molecular Sciences. 2013; 14(7):14346-14373. https://doi.org/10.3390/ijms140714346
Chicago/Turabian StyleRosa, Alessandro, and Ali H. Brivanlou. 2013. "Regulatory Non-Coding RNAs in Pluripotent Stem Cells" International Journal of Molecular Sciences 14, no. 7: 14346-14373. https://doi.org/10.3390/ijms140714346
APA StyleRosa, A., & Brivanlou, A. H. (2013). Regulatory Non-Coding RNAs in Pluripotent Stem Cells. International Journal of Molecular Sciences, 14(7), 14346-14373. https://doi.org/10.3390/ijms140714346






