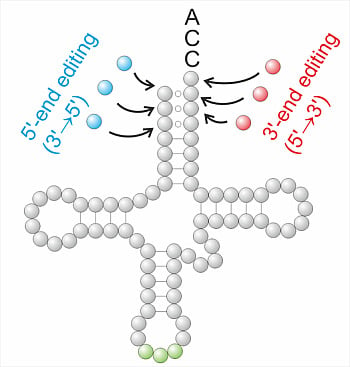
From End to End: tRNA Editing at 5'- and 3'-Terminal Positions
Abstract
:1. Introduction

2. tRNA Editing at 5'-Terminal Positions
2.1. Conventional 5'-End Processing
2.2. Editing of the 5'-End Occurs on a Variety of Eukaryotic tRNAs
2.2.1. Editing of Mismatches at 5'-Terminal Positions

2.2.2. Discovery of Components of the 5'-Editing Enzyme
2.2.3. Nucleases yet to Be Identified
2.2.4. G:U/U:G Base Pair Editing: Diversity among Organisms
2.2.5. Alternative Roles for 5'-End Editing Outside of Mitochondria?
2.2.6. C to U Editing at the 5'-End
2.3. Specialized Editing at the 5'-End of tRNAHis
2.3.1. G-1 Editing of tRNAHis in Eukaryotes

2.3.2. Life without G-1
3. The Heterogeneity of Editing Mechanisms at the tRNA 3'-End
3.1. Template-Independent Mechanisms of tRNA Editing at tRNA 3'-Ends
3.2. Template-Dependent Editing Reactions at tRNA 3'-Ends
4. Saccharomyces cerevisiae as a Model Organism: Evolution of tRNA Editing
4.1. The RNA Surveillance Complex TRAMP4 Shows tRNA 3'-Editing Activity in Yeast
4.2. About the Evolution of tRNA 3'-End Editing

5. Conclusions
| 5'-Editing | 3'-Editing | ||
|---|---|---|---|
| Similarities | Locations | mitochondria and chloroplast in eukaryotes | |
| Mechanism | nucleotide removal and transfer | ||
| Usage | restore base pairs, make functional tRNAs | ||
| Differences | Enzyme | 3'-5'-polymerases (Thg1-like proteins, or TLPs) and unknown enzymes | unknown enzymes, possibly similar to TRAMP complex |
| Nucleotidyl transfer reaction | 3'-5': template-dependent | 5'-3': both template-independent and template-dependent | |
| Substrates | mismatch-containing tRNAs | Overlapping tRNAs and some mismatch-containing tRNAs | |
5.1. What Determines Which Half to Edit?
5.2. Can 5'-Editing Be Used to Process Overlapping tRNAs?
5.3. Editing Machinery and tRNA Surveillance
5.4. Apparent Absence of Editing outside of Eukaryotic Organelles
Author Contributions
Conflicts of Interest
References
- Giegé, R.; Jühling, F.; Pütz, J.; Stadler, P.; Sauter, C.; Florentz, C. Structure of transfer RNAs: Similarity and variability. Wiley Interdiscip. Rev. 2012, 3, 37–61. [Google Scholar] [CrossRef]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Paris, Z.; Fleming Ian, M.C.; Alfonzo, J.D. Determinants of tRNA editing and modification: Avoiding conundrums, affecting function. Semin. Cell Dev. Biol. 2012, 23, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. 2013, 4, 35–48. [Google Scholar] [CrossRef]
- Lohan, A.J.; Gray, M.W. Analysis of 5'- or 3'-terminal tRNA editing: Mitochondrial 5' tRNA editing in Acanthamoeba castellanii as the exemplar. Methods Enzymol. 2007, 424, 223–242. [Google Scholar] [PubMed]
- Gray, M.W. Evolutionary origin of RNA editing. Biochemistry 2012, 51, 5235–5242. [Google Scholar] [CrossRef] [PubMed]
- Schuster, J.; Mörl, M. Mitochondrial tRNA editing. In Mitochondrial Function and Biogenesis; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2004; pp. 81–96. [Google Scholar]
- Zhou, W.; Karcher, D.; Bock, R. Importance of adenosine-to-inosine editing adjacent to the anticodon in an Arabidopsis alanine tRNA under environmental stress. Nucleic Acids Res. 2013, 41, 3362–3372. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Karcher, D.; Bock, R. Identification of enzymes for adenosine-to-inosine editing and discovery of cytidine-to-uridine editing in nucleus-encoded tRNAs of Arabidopsis. Plant Physiol. 2014, 166, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Stanley, B.J.; Kohlway, A.; Mechta, S.; Xiong, Y.; Soll, D. A cytidine deaminase edits C to U in transfer RNAs in archaea. Science 2009, 324, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Marechal-Drouard, L.; Kumar, R.; Remacle, C.; Small, I. RNA editing of larch mitochondrial tRNAHis precursors is a prerequisite for processing. Nucleic Acids Res. 1996, 24, 3229–3234. [Google Scholar] [CrossRef] [PubMed]
- Fey, J.; Weil, J.H.; Tomita, K.; Cosset, A.; Dietrich, A.; Small, I.; Marechal-Drouard, L. Role of editing in plant mitochondrial transfer RNAs. Gene 2002, 286, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Price, D.H.; Gray, M.W. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA 1999, 5, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Mörl, M.; Dörner, M.; Pääbo, S. C to U editing and modifications during the maturation of the mitochondrial tRNAAsp in marsupials. Nucleic Acids Res. 1995, 23, 3380–3384. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, K.M.; Gray, M.W. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science 1993, 259, 812–816. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, K.M.; Gray, M.W. Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res. 1993, 21, 4402. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, J.D.; Blanc, V.; Estevez, A.M.; Rubio, M.A.; Simpson, L. C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999, 18, 7056–7062. [Google Scholar] [CrossRef] [PubMed]
- Janke, A.; Paabo, S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993, 21, 1523–1525. [Google Scholar] [CrossRef] [PubMed]
- Antes, T.; Costandy, H.; Mahendran, R.; Spottswood, M.; Miller, D. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol. Cell. Biol. 1998, 18, 7521–7527. [Google Scholar] [PubMed]
- Walker, S.C.; Engelke, D.R. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 77–102. [Google Scholar] [CrossRef] [PubMed]
- Orellana, O.; Cooley, L.; Soll, D. The additional guanylate at the 5' terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol. Cell. Biol. 1986, 6, 525–529. [Google Scholar] [PubMed]
- Burkard, U.; Soll, D. The 5'-terminal guanylate of chloroplast histidine tRNA is encoded in its gene. J. Biol. Chem. 1988, 263, 9578–9581. [Google Scholar] [PubMed]
- Burkard, U.; Willis, I.; Soll, D. Processing of histidine transfer RNA precursors. Abnormal cleavage site for RNase P. J. Biol. Chem. 1988, 263, 2447–2451. [Google Scholar] [PubMed]
- Kikovska, E.; Brannvall, M.; Kufel, J.; Kirsebom, L.A. Substrate discrimination in RNase P RNA-mediated cleavage: Importance of the structural environment of the RNase P cleavage site. Nucleic Acids Res. 2005, 33, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Kufel, J.; Kirsebom, L.A. Different cleavage sites are aligned differently in the active site of M1 RNA, the catalytic subunit of Escherichia coli RNase P. Proc. Natl. Acad. Sci. USA 1996, 93, 6085–6090. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Singh, D.; Lai, L.B.; Stiffler, M.A.; Lai, H.D.; Foster, M.P.; Gopalan, V. Fidelity of tRNA 5'-maturation: A possible basis for the functional dependence of archaeal and eukaryal RNase P on multiple protein cofactors. Nucleic Acids Res. 2012, 40, 4666–4680. [Google Scholar] [CrossRef] [PubMed]
- Randau, L.; Schroder, I.; Soll, D. Life without RNase P. Nature 2008, 453, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Altman, S. In search of RNase P RNA from microbial genomes. RNA 2004, 10, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Price, D.H.; Gray, M.W. Confirmation of predicted edits and demonstration of unpredicted edits in Acanthamoeba castellanii mitochondrial tRNAs. Curr. Genet. 1999, 35, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Laforest, M.J.; Bullerwell, C.E.; Forget, L.; Lang, B.F. Origin, evolution, and mechanism of 5' tRNA editing in chytridiomycete fungi. RNA 2004, 10, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Bullerwell, C.E. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete Fungus. J. Biol. Chem. 2004, 280, 2463–2470. [Google Scholar] [CrossRef] [PubMed]
- Gott, J.M.; Somerlot, B.H.; Gray, M.W. Two forms of RNA editing are required for tRNA maturation in Physarum mitochondria. RNA 2010, 16, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.G.; Long, Y.; Kinchen, R.D.; Schindel, E.T.; Gray, M.W.; Jackman, J.E. Mitochondrial tRNA 5'-editing in Dictyostelium discoideum and Polysphondylium pallidum. J. Biol. Chem. 2014, 289, 15155–15165. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Hurto, R.L.; Hopper, A.K.; Grayhack, E.J.; Phizicky, E.M. Depletion of Saccharomyces cerevisiae tRNAHis guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m5C. Mol. Cell. Biol. 2005, 25, 8191–8201. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Jackman, J.E.; Lohan, A.J.; Gray, M.W.; Phizicky, E.M. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5' end of tRNAHis. Genes Dev. 2003, 17, 2889–2901. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E. tRNAHis guanylyltransferase catalyzes a 3'-5' polymerization reaction that is distinct from G-1 addition. Proc. Natl. Acad. Sci. USA 2006, 103, 8640–8645. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.A.; Jackman, J.E. Kinetic analysis of 3'-5' nucleotide addition catalyzed by eukaryotic tRNAHis guanylyltransferase. Biochemistry 2012, 51, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.A.; Jackman, J.E. Saccharomyces cerevisiae Thg1 uses 5'-pyrophosphate removal to control addition of nucleotides to tRNAHis. Biochemistry 2014, 53, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Gott, J.M.; Gray, M.W. Doing it in reverse: 3'-to-5' polymerization by the Thg1 superfamily. RNA 2012, 18, 886–899. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.G.; Rao, B.S.; Jackman, J.E. Template-dependent 3'-5' nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc. Natl. Acad. Sci. USA 2010, 107, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, I.U.; Randau, L.; Tomko, R.J., Jr.; Söll, D. 3'–5' tRNAHis guanylyltransferase in bacteria. FEBS Lett. 2010, 584, 3567–3572. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.S.; Maris, E.L.; Jackman, J.E. tRNA 5'-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from bacteria and archaea. Nucleic Acids Res. 2011, 39, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.G.; Long, Y.; Willcox, A.; Gott, J.M.; Gray, M.W.; Jackman, J.E. A role for tRNAHis guanylyltransferase (Thg1)-like proteins from Dictyostelium discoideum in mitochondrial 5'-tRNA editing. RNA 2011, 17, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.S.; Mohammad, F.; Gray, M.W.; Jackman, J.E. Absence of a universal element for tRNAHis identity in Acanthamoeba castellanii. Nucleic Acids Res. 2013, 41, 1885–1894. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Altman, S. Substrate recognition by human RNase P: Identification of small, model substrates for the enzyme. EMBO J. 1995, 14, 159–168. [Google Scholar] [PubMed]
- Dewe, J.M.; Whipple, J.M.; Chernyakov, I.; Jaramillo, L.N.; Phizicky, E.M. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 2012, 18, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Whipple, J.M.; Lane, E.A.; Chernyakov, I.; D’Silva, S.; Phizicky, E.M. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011, 25, 1173–1184. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, M.; Sekine, S.; Chong, Y.E.; Guo, M.; Yang, X.L.; Gamper, H.; Hou, Y.M.; Schimmel, P.; Yokoyama, S. The selective tRNA aminoacylation mechanism based on a single G·U pair. Nature 2014, 510, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, K.; Schneider, J.; McClain, W.H. Functional evidence for indirect recognition of G·U in tRNAAla by alanyl-tRNA synthetase. Science 1996, 271, 195–197. [Google Scholar] [CrossRef] [PubMed]
- McClain, W.H.; Gabriel, K.; Schneider, J. Specific function of a G·U wobble pair from an adjacent helical site in tRNAAla during recognition by alanyl-tRNA synthetase. RNA 1996, 2, 105–109. [Google Scholar]
- Lizano, E.; Scheibe, M.; Rammelt, C.; Betat, H.; Mörl, M. A comparative analysis of CCA-adding enzymes from human and E. coli: Differences in CCA addition and tRNA 3'-end repair. Biochimie 2008, 90, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Deutscher, M.P. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 1987, 6, 2473–2477. [Google Scholar] [PubMed]
- Binder, S.; Marchfelder, A.; Brennicke, A. RNA editing of tRNAPhe and tRNACys in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet. 1994, 244, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Marechal-Drouard, L.; Cosset, A.; Remacle, C.; Ramamonjisoa, D.; Dietrich, A. A single editing event is a prerequisite for efficient processing of potato mitochondrial phenylalanine tRNA. Mol. Cell. Biol. 1996, 16, 3504–3510. [Google Scholar] [PubMed]
- Himeno, H.; Hasegawa, T.; Ueda, T.; Watanabe, K.; Miura, K.; Shimizu, M. Role of the extra G-C pair at the end of the acceptor stem of tRNAHis in aminoacylation. Nucleic Acids Res. 1989, 17, 7855–7863. [Google Scholar] [CrossRef] [PubMed]
- Nameki, N.; Asahara, H.; Shimizu, M.; Okada, N.; Himeno, H. Identity elements of Saccharomyces cerevisiae tRNAHis. Nucleic Acids Res. 1995, 23, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Rosen, A.E.; Musier-Forsyth, K. Recognition of G-1: C73 atomic groups by Escherichia coli histidyl-tRNA synthetase. J. Am. Chem. Soc. 2004, 126, 64–65. [Google Scholar] [CrossRef] [PubMed]
- Giege, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Francklyn, C. Cytosine 73 is a discriminator nucleotide in vivo for histidyl-tRNA in Escherichia coli. J. Biol. Chem. 1994, 269, 10022–10027. [Google Scholar] [PubMed]
- Preston, M.A.; Phizicky, E.M. The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by over-expression of tRNAHis and its synthetase. RNA 2010, 16, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Cooley, L.; Appel, B.; Söll, D. Post-transcriptional nucleotide addition is responsible for the formation of the 5' terminus of histidine tRNA. Proc. Natl. Acad. Sci. USA 1982, 79, 6475–6479. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sobral, B.W.; Williams, K.P. Loss of a universal tRNA feature. J. Bacteriol. 2007, 189, 1954–1962. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Gogakos, T.; Babina, A.M.; Soll, D.; Randau, L. Change of tRNA identity leads to a divergent orthogonal histidyl-tRNA synthetase/tRNAHis pair. Nucleic Acids Res. 2011, 39, 2286–2293. [Google Scholar] [CrossRef] [PubMed]
- Ardell, D.H.; Andersson, S.G. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006, 34, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.; Lang, B.F. Mitochondrial 3' tRNA editing in the jakobid Seculamonas ecuadoriensis: A novel mechanism and implications for tRNA processing. RNA 2004, 10, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Reichert, A.; Rothbauer, U.; Morl, M. Processing and editing of overlapping tRNAs in human mitochondria. J. Biol. Chem. 1998, 273, 31977–31984. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, S.; Paabo, S. Transfer RNA editing in land snail mitochondria. Proc. Natl. Acad. Sci. USA 1995, 92, 10432–10435. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, S.; Paabo, S. Polyadenylation creates the discriminator nucleotide of chicken mitochondrial tRNATyr. J. Mol. Biol. 1997, 265, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, S.I.; Paabo, S. tRNA editing in metazoans. Nature 1995, 377, 490. [Google Scholar] [CrossRef] [PubMed]
- Kurland, C.G. Evolution of mitochondrial genomes and the genetic code. Bioessays 1992, 14, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Ueda, T.; Watanabe, K. RNA editing in the acceptor stem of squid mitochondrial tRNATyr. Nucleic Acids Res. 1996, 24, 4987–4991. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, S.; Misch, A.; Sprinzl, M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic. Acids Res. 1993, 21, 3011–3015. [Google Scholar] [CrossRef] [PubMed]
- Nagaike, T.; Suzuki, T.; Ueda, T. Polyadenylation in mammalian mitochondria: Insights from recent studies. Biochim. Biophys. Acta 2008, 1779, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.H.; Tong, L. Mitochondrial poly(A) polymerase and polyadenylation. Biochim. Biophys. Acta 2012, 1819, 992–997. [Google Scholar] [CrossRef] [PubMed]
- Rorbach, J.; Nicholls, T.J.; Minczuk, M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res. 2011, 39, 7750–7763. [Google Scholar] [CrossRef] [PubMed]
- Reichert, A.S.; Mörl, M. Repair of tRNAs in metazoan mitochondria. Nucleic Acids Res. 2000, 28, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Schuster, J.; Betat, H.; Morl, M. Is yeast on its way to evolving tRNA editing? EMBO Rep. 2005, 6, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Cole, T.E.; Brasier, C.M.; Buck, K.W. Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch Elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 1998, 246, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, I.E. Mitochondria; Wiley-Liss: New York, NY, USA, 1999. [Google Scholar]
- Li, Z.; Deutscher, M.P. The role of individual exoribonucleases in processing at the 3' end of Escherichia coli tRNA precursors. J. Biol. Chem. 1994, 269, 6064–6071. [Google Scholar] [PubMed]
- Li, Z.; Deutscher, M.P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA 2002, 8, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Ow, M.C.; Kushner, S.R. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 2002, 16, 1102–1115. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wang, X.; Liu, F.; Guenther, U.-P.; Srinivasan, S.; Anderson, J.T.; Jankowsky, E. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell 2011, 145, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, H.; Tretbar, S.; Betat, H.; Morl, M. The TRAMP complex shows tRNA editing activity in S. cerevisiae. Mol. Biol. Evol. 2012, 29, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Vanácová, S.; Wolf, J.; Martin, G.; Blank, D.; Dettwiler, S.; Friedlein, A.; Langen, H.; Keith, G.; Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005, 3, e189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyers, F.; Rougemaille, M.; Badis, G.; Rousselle, J.-C.; Dufour, M.-E.; Boulay, J.; Regnault, B.; Devaux, F.; Namane, A.; Seraphin, B.; et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 2005, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Cell and genome coevolution: Facultative anaerobiosis, glycosomes and kinetoplastan RNA editing. Trends Genet. 1997, 13, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Covello, P.S.; Gray, M.W. On the evolution of RNA editing. Trends Genet. 1993, 9, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Khersonsky, O.; Tawfik, D.S. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010, 79, 471–505. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.; Thiemann, O.H.; Savill, N.J.; Alfonzo, J.D.; Maslov, D.A. Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. USA 2000, 97, 6986–6993. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betat, H.; Long, Y.; Jackman, J.E.; Mörl, M. From End to End: tRNA Editing at 5'- and 3'-Terminal Positions. Int. J. Mol. Sci. 2014, 15, 23975-23998. https://doi.org/10.3390/ijms151223975
Betat H, Long Y, Jackman JE, Mörl M. From End to End: tRNA Editing at 5'- and 3'-Terminal Positions. International Journal of Molecular Sciences. 2014; 15(12):23975-23998. https://doi.org/10.3390/ijms151223975
Chicago/Turabian StyleBetat, Heike, Yicheng Long, Jane E. Jackman, and Mario Mörl. 2014. "From End to End: tRNA Editing at 5'- and 3'-Terminal Positions" International Journal of Molecular Sciences 15, no. 12: 23975-23998. https://doi.org/10.3390/ijms151223975
APA StyleBetat, H., Long, Y., Jackman, J. E., & Mörl, M. (2014). From End to End: tRNA Editing at 5'- and 3'-Terminal Positions. International Journal of Molecular Sciences, 15(12), 23975-23998. https://doi.org/10.3390/ijms151223975