Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken Myoblast Fusion
Abstract
:1. Introduction
2. Results
2.1. cDNA Sequence, Genomic Structure and Protein Conservation of the Chicken Myomaker Gene
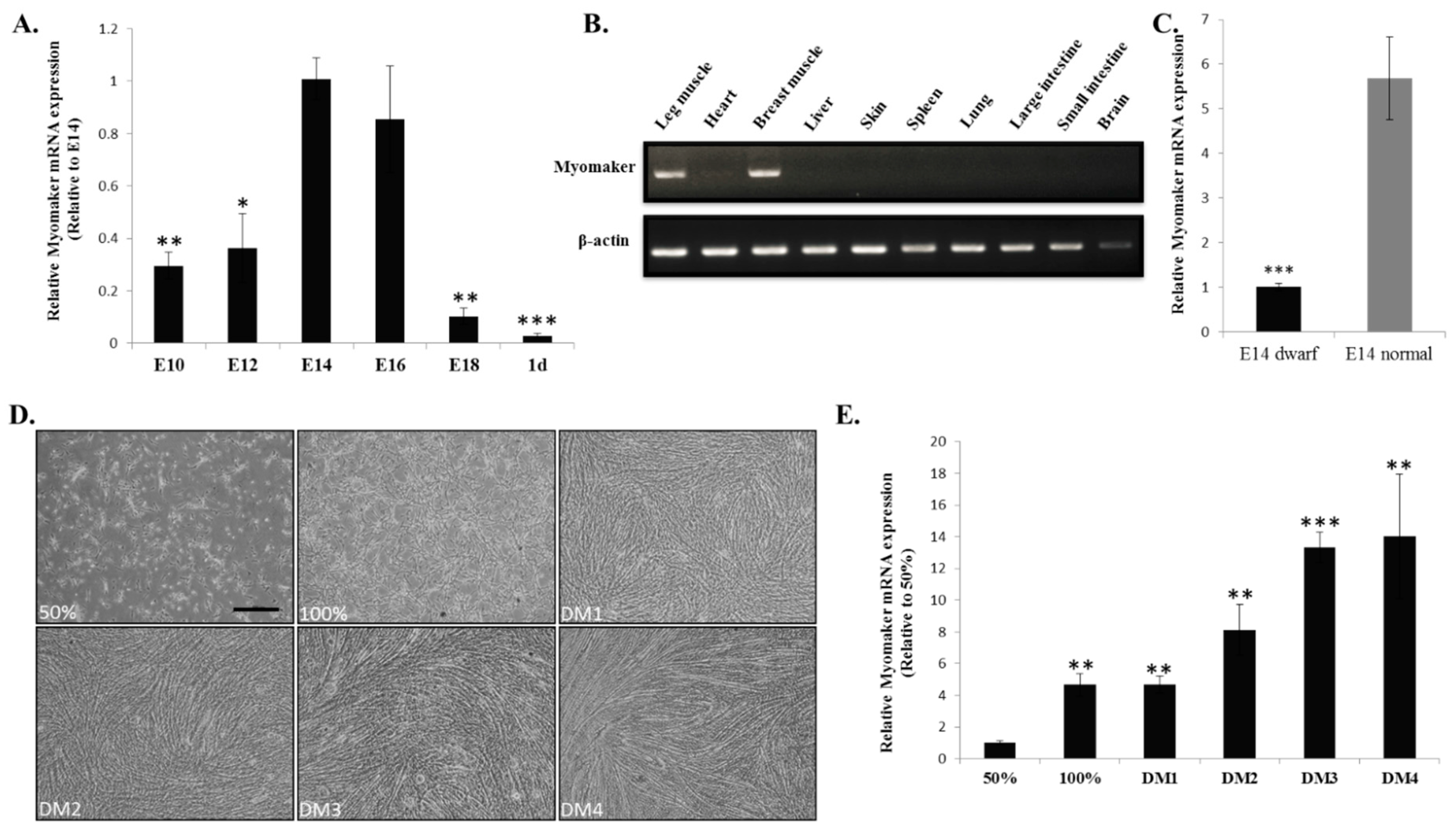
2.2. Myomaker mRNA Expression during Chicken Skeletal Muscle Development

2.3. Myomaker Is Essential for Myoblast Fusion in Chicken

2.4. Myomaker Transcription Is Controlled by a Conserved Promoter E-Box Element in Chicken

2.5. MYOD and MYOG Bind to the E-Box 1 Region and Regulate Myomaker Transcription

2.6. miR-140-3p Binds Directly to the 3ʹ UTR of Myomaker and Inhibits Myomaker Expression in Vitro
3. Discussion

4. Experimental Section
4.1. Animals and Cells
4.2. cDNA Synthesis and Quantitative Real-Time PCR (qPCR)
4.3. The 5ʹ and 3ʹ Rapid Amplification of cDNA Ends (RACE)
4.4. Immunofluorescence
4.5. ChIP Assays
4.6. Transfections
4.7. Plasmid Construction
4.7.1. pcDNA-3.1 Gene Overexpression Vectors
4.7.2. pmirGLO Dual-Luciferase Reporters
4.7.3. Myomaker Promoter Reporter Plasmid
4.8. Target Prediction
4.9. Dual Luciferase Reporter Assay
4.10. Cell-Cycle Analysis
4.11. Statistical Analysis
4.12. Ethics Standards
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pavlath, G.K.; Horsley, V. Cell fusion in skeletal muscle—Central role of NFATC2 in regulating muscle cell size. Cell Cycle 2003, 2, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Abmayr, S.M.; Balagopalan, L.; Galletta, B.J.; Hong, S.J. Cell and molecular biology of myoblast fusion. Int. Rev. Cytol. 2003, 225, 33–89. [Google Scholar] [PubMed]
- Abmayr, S.M.; Pavlath, G.K. Myoblast fusion: lessons from flies and mice. Development 2012, 139, 641–656. [Google Scholar] [CrossRef] [PubMed]
- Rochlin, K.; Yu, S.; Roy, S.; Baylies, M.K. Myoblast fusion: When it takes more to make one. Dev. Biol. 2010, 341, 66–83. [Google Scholar] [CrossRef] [PubMed]
- Doherty, K.R.; Cave, A.; Davis, D.B.; Delmonte, A.J.; Posey, A.; Earley, J.U.; Hadhazy, M.; McNally, E.M. Normal myoblast fusion requires myoferlin. Development 2005, 132, 5565–5575. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.M.; Pavlath, G.K. Mannose receptor regulates myoblast motility and muscle growth. J. Cell Biol. 2006, 174, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gomez, M.; Coutts, N.; Suster, M.L.; Landgraf, M.; Bate, M. Myoblasts incompetent encodes a Zinc finger transcription factor required to specify fusion-competent myoblasts in Drosophila. Development 2002, 129, 133–141. [Google Scholar] [PubMed]
- Strunkelnberg, M.; Bonengel, B.; Moda, L.M.; Hertenstein, A.; de Couet, H.G.; Ramos, R.G.; Fischbach, K.F. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development 2001, 128, 4229–4239. [Google Scholar] [PubMed]
- Artero, R.D.; Castanon, I.; Baylies, M.K. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development 2001, 128, 4251–4464. [Google Scholar] [PubMed]
- Bour, B.A.; Chakravarti, M.; West, J.M.; Abmayr, S.M. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000, 14, 1498–1511. [Google Scholar] [PubMed]
- Millay, D.P.; O’Rourke, J.R.; Sutherland, L.B.; Bezprozvannaya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 2013, 499, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Millay, D.P.; Sutherland, L.B.; Bassel-Duby, R.; Olson, E.N. Myomaker is essential for muscle regeneration. Genes Dev. 2014, 28, 1641–1646. [Google Scholar] [CrossRef] [PubMed]
- Landemaine, A.; Rescan, P.Y.; Gabillard, J.C. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem. Biophys. Res. Commun. 2014, 451, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011, 12, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Berkes, C.A.; Tapscott, S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Buschhausen-Denker, G.; Bober, E.; Tannich, E.; Arnold, H.H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989, 8, 701–709. [Google Scholar] [PubMed]
- Edmondson, D.G.; Olson, E.N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1990, 4, 1450. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Hasty, P.; Bradley, A.; Morris, J.H.; Edmondson, D.G.; Venuti, J.M.; Olson, E.N.; Klein, W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993, 364, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Kumar, R.M.; Penn, B.H.; Berkes, C.A.; Kooperberg, C.; Boyer, L.A.; Young, R.A.; Tapscott, S.J. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006, 25, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wu, H.; Ye, Y.; Li, Z.; Hao, S.; Kong, L.; Zheng, X.; Lin, S.; Nie, Q.; Zhang, X. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014, 5, e1347. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.K.; Cogburn, L.A.; Burnside, J. Dysfunctional growth hormone receptor in a strain of sex-linked dwarf chicken: Evidence for a mutation in the intracellular domain. J. Endocrinol. 1994, 142, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Kassar-Duchossoy, L.; Gayraud-Morel, B.; Gomes, D.; Rocancourt, D.; Buckingham, M.; Shinin, V.; Tajbakhsh, S. Mrf4 determines skeletal muscle identity in Myf5: Myod double-mutant mice. Nature 2004, 431, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Blais, A.; Tsikitis, M.; Acosta-Alvear, D.; Sharan, R.; Kluger, Y.; Dynlacht, B.D. An initial blueprint for myogenic differentiation. Genes Dev. 2005, 19, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Nie, Q.; Zhang, X. MicroRNAs involved in skeletal muscle differentiation. J. Genet. Genom. 2013, 40, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Liu, N.; Olson, E.N. MicroRNAs flex their muscles. Trends Genet. 2008, 24, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Frasch, M. MicroRNAs in muscle differentiation: lessons from Drosophila and beyond. Curr. Opin. Genet. Dev. 2006, 16, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, T.A.; Jakobsen, R.B.; Mikkelsen, T.S.; Brinchmann, J.E. microRNA-140 targets RALA and regulates chondrogenic differentiation of human mesenchymal stem cells by translational enhancement of SOX9 and ACAN. Stem Cells Dev. 2014, 23, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Rakoczy, J.; Fernandez-Valverde, S.L.; Glazov, E.A.; Wainwright, E.N.; Sato, T.; Takada, S.; Combes, A.N.; Korbie, D.J.; Miller, D.; Grimmond, S.M.; et al. MicroRNAs-140–5p/140–3p modulate Leydig cell numbers in the developing mouse testis. Biol. Reprod. 2013, 88, 143. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.X.; Huang, W.; Wang, X.B.; Lv, G.H.; Li, J.; Deng, Y.W. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4877–4885. [Google Scholar] [PubMed]
- Zhu, X.; Chen, D.; Hu, Y.; Wu, P.; Wang, K.; Zhang, J.; Chu, W.; Zhang, J. The microRNA Signature in Response to Nutrient Restriction and Refeeding in Skeletal Muscle of Chinese Perch (Siniperca chuatsi). Mar. Biotechnol. 2015, 17, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Grobet, L.; Martin, L.J.; Poncelet, D.; Pirottin, D.; Brouwers, B.; Riquet, J.; Schoeberlein, A.; Dunner, S.; Menissier, F.; Massabanda, J.; et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 1997, 17, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Langley, B.; Berry, C.; Sharma, M.; Kirk, S.; Bass, J.; Kambadur, R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000, 275, 40235–40243. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J. Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 2004, 20, 61–86. [Google Scholar] [CrossRef] [PubMed]
- Jude, J.A.; Dileepan, M.; Subramanian, S.; Solway, J.; Panettieri, R.J.; Walseth, T.F.; Kannan, M.S. miR-140-3p regulation of TNFα-induced CD38 expression in human airway smooth muscle cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2012, 303, L460–L468. [Google Scholar] [CrossRef] [PubMed]
- Keren, A.; Tamir, Y.; Bengal, E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 2006, 252, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Zetser, A.; Gredinger, E.; Bengal, E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem. 1999, 274, 5193–5200. [Google Scholar] [CrossRef] [PubMed]
- Bakkar, N.; Guttridge, D.C. NF-κB signaling: A tale of two pathways in skeletal myogenesis. Physiol. Rev. 2010, 90, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Bakkar, N.; Wang, J.; Ladner, K.J.; Wang, H.; Dahlman, J.M.; Carathers, M.; Acharyya, S.; Rudnicki, M.A.; Hollenbach, A.D.; Guttridge, D.C. IKK/NF-κB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J. Cell Biol. 2008, 180, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.B.; Xu, H.; Xie, S.J.; Zhou, H.; Qu, L.H. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS ONE 2011, 6, e29173. [Google Scholar] [CrossRef] [PubMed]
- Boutz, P.L.; Chawla, G.; Stoilov, P.; Black, D.L. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007, 21, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, W.; Li, E.; Nie, Q.; Zhang, X. Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken Myoblast Fusion. Int. J. Mol. Sci. 2015, 16, 26186-26201. https://doi.org/10.3390/ijms161125946
Luo W, Li E, Nie Q, Zhang X. Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken Myoblast Fusion. International Journal of Molecular Sciences. 2015; 16(11):26186-26201. https://doi.org/10.3390/ijms161125946
Chicago/Turabian StyleLuo, Wen, Erxin Li, Qinghua Nie, and Xiquan Zhang. 2015. "Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken Myoblast Fusion" International Journal of Molecular Sciences 16, no. 11: 26186-26201. https://doi.org/10.3390/ijms161125946
APA StyleLuo, W., Li, E., Nie, Q., & Zhang, X. (2015). Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken Myoblast Fusion. International Journal of Molecular Sciences, 16(11), 26186-26201. https://doi.org/10.3390/ijms161125946






