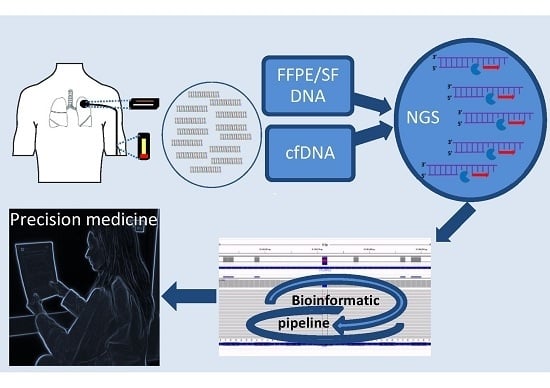
Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGM™ Platform
Abstract
:1. Introduction
2. Results
2.1. Assessment of NGS (Next-Generation Sequencing) Detection Limit, Specificity, Sensitivity, and Positive Predictive Value
2.2. FFPE (Formalin-Fixed Paraffin-Embedded) Quality Control
2.3. FFPE and Snap Frozen (SF) NGS
| Patient | Tumor Type | EGFR and KRAS Status by Sanger Sequencing | EGFR and KRAS Status Confirmed by VC Software | Allele Name |
|---|---|---|---|---|
| 1 | SF | NM_033360.3 (KRAS): c.34G>C | Confirmed KRAS mutation | COSM518 |
| (p.Gly12Arg) | AF: 29.8% | |||
| 1 | FFPE | NM_033360.3 (KRAS): c.34G>C | Confirmed KRAS mutation | COSM518 |
| (p.Gly12Arg) | AF: 58.6% | |||
| 2 | SF | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 24.1% | |||
| 2 | FFPE | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 6.4% | |||
| 3 | SF | NM_005228.3 (EGFR): c.2239_2240TT>CC | Confirmed EGFR mutation | COSM24267 |
| (p.Lys747Pro) | AF: 37.8% | |||
| 3 | FFPE | NM_005228.3 (EGFR): c.2239_2240TT>CC | Confirmed EGFR mutation | COSM24267 |
| (p.Lys747Pro) | AF: 45.6% | |||
| 4 | SF | NM_033360.3 (KRAS): c.35G>T | Confirmed KRAS mutation | COSM520 |
| (p.Gly12Val) | AF: 24.0% | |||
| 4 | FFPE | NM_033360.3 (KRAS): c.35G>T | Confirmed KRAS mutation | COSM520 |
| (p.Gly12Val) | AF: 23.2% | |||
| 5 | SF | NM_005228.3 (EGFR): c.2237_2255delinsT | Confirmed EGFR mutation | COSM12384 |
| (p.Glu746_Ser752delinsVal) | AF: 5.6% | |||
| 5 | FFPE | NM_005228.3 (EGFR): c.2237_2255delinsT | Confirmed EGFR mutation | COSM12384 |
| (p.Glu746_Ser752delinsVal) | AF: 10.5% | |||
| 6 | SF | NM_005228.3 (EGFR): c.2236_2250del | Confirmed EGFR mutation | COSM6225 |
| (p.Glu746_Ala750del) | AF: 7.8% | |||
| 6 | FFPE | NM_005228.3 (EGFR): c.2236_2250del | Confirmed EGFR mutation | COSM6225 |
| (p.Glu746_Ala750del) | AF: 45.8% | |||
| 7 | SF | NM_033360.3 (KRAS): c.35G>A | Confirmed KRAS mutation | COSM521 |
| (p.Gly12Asp) | AF: 38.6% | |||
| 7 | FFPE | NM_033360.3 (KRAS): c.35G>A | Confirmed KRAS mutation | COSM521 |
| (p.Gly12Asp) | AF: 26.3% | |||
| 8 | SF | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 50.5% | |||
| 8 | FFPE | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 24.5% | |||
| 9 | SF | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 42.0% | |||
| 9 | FFPE | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 41.7% | |||
| 10 | SF | NM_005228.3 (EGFR): c.2573T>G | Confirmed EGFR mutation | COSM6224 |
| (p.Leu858Arg) | AF: 45.3% | |||
| 10 | FFPE | NM_005228.3 (EGFR): c.2573T>G | Confirmed EGFR mutation | COSM6224 |
| (p.Leu858Arg) | AF: 40.2% | |||
| 11 | SF | NM_033360.3 (KRAS): c.37G>T | Confirmed KRAS mutation | COSM527 |
| (p.Gly13Cys) | AF: 32.0% | |||
| 11 | FFPE | NM_033360.3 (KRAS): c.37G>T | Confirmed KRAS mutation | COSM527 |
| (p.Gly13Cys) | AF: 24.2% | |||
| 12 | SF | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 31.0% | |||
| 12 | FFPE | NM_033360.3 (KRAS): c.34G>T | Confirmed KRAS mutation | COSM516 |
| (p.Gly12Cys) | AF: 42.6% |

2.4. cfDNA NGS
3. Discussion

4. Experimental Section
4.1. Sample Collection
| Patient | Block Age | % TC | SF | FFPE | cfDNA | Histology | TNM | Stage | Age at Diagnosis | G | Smoking Habits | PS | OS | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2012 | >50 | x | x | x | SCC | T1b, N0, M0 | IA | 77 | M | S | 1 | 30 | A |
| 2 | 2010 | >50 | x | x | x | ADK | T1a,N2,M0 | IIIA | 67 | M | NS | 1 | 18 | DOD |
| 3 | 2013 | >50 | x | x | x | ADK | T2, N2, M1a | IV | 76 | F | NS | 1 | 14 | DOD |
| 4 | 2010 | >50 | x | x | x | ADK | T2b,N0,MX | IIA | 76 | M | FS | 1 | 11 | DOD |
| 5 | 2013 | >50 | x | x | NA | ADK | T2a,N2,M1b | IV | 75 | F | NS | 1 | 13 | A |
| 6 | 2011 | >50 | x | x | NA | ADK | T2a, N0, MX | IB | 78 | F | NS | 1 | 29 | DOD |
| 7 | 2013 | >50 | x | x | x | ADK | T2a, N2, M1b | IV | 65 | M | S | 1 | 3 | DOD |
| 8 | 2013 | >50 | x | x | x | ADK | T2b, N1, M0 | IIB | 59 | F | S | 1 | 23 | A |
| 9 | 2012 | >50 | x | x | x | ADK | T2a, N0, MX | IB | 72 | M | FS | 0 | 28 | A |
| 10 | 2011 | >50 | x | x | NA | ADK | T2b, N0, M0 | IIB | 66 | F | FS | 1 | 42 | A |
| 11 | 2010 | >50 | x | x | x | ADK | T1a, N0, M0 | IA | 77 | M | FS | 0 | 49 | DOD |
| 12 | 2010 | >50 | x | x | x | ADK | T1a, N0, M0 | IA | 60 | F | S | 1 | 32 | DOD |
| Patient | Block Age | % TC | SF | FFPE | cfDNA | Histology | TNM | Stage | Age at Diagnosis | G | Smoking Habits | PS | OS | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 2015 | >50 | NA | x | NA | ADK | T2, N2, M0 | IIIB | 59 | M | S | 0 | 5 | A |
| 14 | 2014 | >50 | NA | x | NA | ADK | Tx, Nx, M1a | IV | 42 | M | NS | 0 | 14 | A |
| 15 | 2015 | >50 | NA | x | NA | ADK | T3, N3, M1a | IV | 64 | M | FS | 1 | 4 | A |
| 16 | 2014 | >50 | NA | x | NA | ADK | T4, N2, M1b | IV | 77 | F | NS | 0 | 12 | A |
| 17 | 2014 | >50 | NA | x | NA | ADK | T3, N2, M0 | IIIA | 70 | F | NS | 1 | 8 | A |
| 18 | 2012 | >50 | NA | x | NA | ADK | T3, N2, M1b | IV | 63 | M | S | 1 | 37 | A |
| 19 | 2014 | >50 | NA | x | NA | ADK | T4, N2, M1b | IV | 70 | M | FS | 1 | 2 | DOD |
| 20 | 2014 | >50 | NA | x | NA | ADK | T4, N0, M0 | IIIA | 68 | M | FS | 0 | 16 | A |
| 21 | 2011 | >50 | NA | x | NA | ADK | T1a, N0, M0 | IA | 71 | F | NS | 1 | 42 | DOD |
| 22 | 2014 | >50 | NA | x | NA | ADK | T2, N2, M1b | IV | 81 | F | NS | 1 | 10 | DOD |
| 23 | 2013 | >50 | NA | x | NA | ADK | T1b, N2, M0 | IIIA | 72 | M | FS | 1 | 22 | A |
| 24 | 2014 | >50 | NA | x | NA | ADK | T3, N3, M1b | IV | 61 | M | NS | 1 | 11 | DOD |
| 25 | 2015 | >50 | NA | x | NA | ADK | Tx, Nx, M1b | IV | 55 | F | FS | 0 | 7 | A |
4.2. DNA Extraction and Quality Control
4.3. Whole Genome Amplification (WGA)
4.4. Next Generation Sequencing (NGS)
4.5. NGS Data Analysis
4.6. NGS Parameter Performance
4.7. Sanger Sequencing (SS) and Pyrosequencing (PS)
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Miller, V.; Zakowski, M.; Doherty, J.; Politi, K.; Sarkaria, I.; Singh, B.; Heelan, R.; Rusch, V.; Fulton, L.; et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA 2004, 101, 13306–13311. [Google Scholar] [CrossRef] [PubMed]
- Coco, S.; Truini, A.; Vanni, I.; dal Bello, M.G.; Alama, A.; Rijavec, E.; Genova, C.; Barletta, G.; Sini, C.; Burrafato, G.; et al. Next generation sequencing in non-small cell lung cancer: New avenues toward the personalized medicine. Curr. Drug Targets 2015, 16, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Biotechnology 1993, 11, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Ronaghi, M.; Uhlén, M.; Nyrén, P. A sequencing method based on real-time pyrophosphate. Science 1998, 281, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Hantson, I.; Dooms, C.; Verbeken, E.; Vandenberghe, P.; Vliegen, L.; Roskams, T.; Borght, S.V.; Nackaerts, K.; Wauters, I.; Vansteenkiste, J. Performance of standard procedures in detection of EGFR mutations in daily practice in advanced NSCLC patients selected according to the ESMO guideline: A large Caucasian cohort study. Transl. Respir. Med. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Cronin, M.; Ross, J.S. Comprehensive next-generation cancer genome sequencing in the era of targeted therapy and personalized oncology. Biomark. Med. 2011, 5, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Couraud, S.; Vaca-Paniagua, F.; Villar, S.; Oliver, J.; Schuster, T.; Blanché, H.; Girard, N.; Trédaniel, J.; Guilleminault, L.; Gervais, R.; et al. BioCAST/IFCT-1002 investigators: Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin. Cancer Res. 2014, 20, 4613–4624. [Google Scholar] [CrossRef] [PubMed]
- Nomenclature for the description of sequence variants. Available online: http://www.hgvs.org/mutnomen/ (accessed on 10 April 2015).
- Den Dunnen, J.T.; Antonarakis, S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: A discussion. Hum. Mutat. 2000, 15, 7–12. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- COSMIC (Catalogue of Somatic Mutations in Cancer). Available online: http://cancer.sanger.ac.uk/cosmic (accessed on 28 February 2015).
- Choudhary, A.; Mambo, E.; Sanford, T.; Boedigheimer, M.; Twomey, B.; Califano, J.; Hadd, A.; Oliner, K.S.; Beaudenon, S.; Latham, G.J.; et al. Evaluation of an integrated clinical workflow for targeted next-generation sequencing of low-quality tumor DNA using a 51-gene enrichment panel. BMC Med. Genom. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.; Sah, S.; Latham, G.J.; Patel, R.; Song, Q.; Koeppen, H.; Tam, R.; Schleifman, E.; Mashhedi, H.; et al. Profiling cancer gene mutations in clinical formalin-fixed, paraffin-embedded colorectal tumor specimens using targeted next-generation sequencing. Oncologist 2014, 19, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Hadd, A.G.; Houghton, J.; Choudhary, A.; Sah, S.; Chen, L.; Marko, A.C.; Sanford, T.; Buddavarapu, K.; Krosting, J.; Garmirek, L.; et al. Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J. Mol. Diagn. 2013, 15, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, I.S.; Devarakonda, S.; Lockwood, C.M.; Spencer, D.H.; Guebert, K.; Bredemeyer, A.J.; al-Kateb, H.; Nguyen, T.T.; Duncavage, E.J.; Cottrell, C.E.; et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015, 121, 631–639. [Google Scholar] [CrossRef] [PubMed]
- D’Haene, N.; le Mercier, M.; de Nève, N.; Blanchard, O.; Delaunoy, M.; el Housni, H.; Dessars, B.; Heimann, P.; Remmelink, M.; Demetter, P.; et al. Clinical Validation of Targeted Next Generation Sequencing for Colon and Lung Cancers. PLoS ONE 2015, 10, e0138245. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.; Chen, L.; Houghton, J.; Kemppainen, J.; Marko, A.C.; Zeigler, R.; Latham, G.J. Functional DNA quantification guides accurate next-generation sequencing mutation detection in formalin-fixed, paraffin-embedded tumor biopsies. Genome Med. 2013, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Simbolo, M.; Gottardi, M.; Corbo, V.; Fassan, M.; Mafficini, A.; Malpeli, G.; Lawlor, R.T.; Scarpa, A. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS ONE 2013, 8, e62692. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.G.; Russ, C.; Costello, M.; Hollinger, A.; Lennon, N.J.; Hegarty, R.; Nusbaum, C.; Jaffe, D.B. Characterizing and measuring bias in sequence data. Genome Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Wilm, A.; Aw, P.P.; Bertrand, D.; Yeo, G.H.; Ong, S.H.; Wong, C.H.; Khor, C.C.; Petric, R.; Hibberd, M.L.; Nagarajan, N. LoFreq: A sequence quality aware, ultra-sensitive variant caller for uncovering cell population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef] [PubMed]
- Genome Analysis Toolkit (GATK). Available online: https://www.broadinstitute.org/gatk/index.php (accessed on 3 February 2015).
- Boland, J.F.; Chung, C.C.; Roberson, D.; Mitchell, J.; Zhang, X.; Im, K.M.; He, J.; Chanock, S.J.; Yeager, M.; Dean, M. The new sequencer on the block: Comparison of Life Technology’s Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Hum. Genet. 2013, 132, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Sakurada, A.; Lara-Guerra, H.; Liu, N.; Shepherd, F.A.; Tsao, M.S. Tissue heterogeneity of EGFR mutation in lung adenocarcinoma. J. Thorac. Oncol. 2008, 3, 527–529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fujimoto, J.; Zhang, J.; Wedge, D.C.; Song, X.; Zhang, J.; Seth, S.; Chow, C.W.; Cao, Y.; Gumbs, C.; et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014, 346, 256–259. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, E.C.; McGranahan, N.; Mitter, R.; Salm, M.; Wedge, D.C.; Yates, L.; Jamal-Hanjani, M.; Shafi, S.; Murugaesu, N.; Rowan, A.J.; et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014, 346, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Berger, A.H.; Hammerman, P.S.; Hernandez, B.; Pugh, T.J.; Hodis, E.; Cho, J.; Suh, J.; Capelletti, M.; Sivachenko, A.; et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012, 150, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar]
- Zhang, X.; Miao, X.; Guo, Y.; Tan, W.; Zhou, Y.; Sun, T.; Wang, Y.; Lin, D. Genetic polymorphisms in cell cycle regulatory genes MDM2 and TP53 are associated with susceptibility to lung cancer. Hum. Mutat. 2006, 27, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Chen, Y.; Gao, C.; Wei, Y.; Cao, G.; Lu, N.; Hou, Y.; Jiang, X.; Wang, J. Polymorphisms of p53 and MDM2 genes are associated with severe toxicities in patients with non-small cell lung cancer. Cancer Biol. Ther. 2014, 15, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Govindan, R.; Ding, L.; Griffith, M.; Subramanian, J.; Dees, N.D.; Kanchi, K.L.; Maher, C.A.; Fulton, R.; Fulton, L.; Wallis, J.; et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012, 50, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cui, X.; Schmitt, K.; Hubert, R.; Navidi, W.; Arnheim, N. Whole genome amplification from a single cell: Implications for genetic analysis. Proc. Natl. Acad. Sci. USA 1992, 89, 5847–5851. [Google Scholar] [CrossRef] [PubMed]
- Telenius, H.; Carter, N.P.; Bebb, C.E.; Nordenskjold, M.; Ponder, B.A.; Tunnacliffe, A. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics 1992, 13, 718–725. [Google Scholar] [CrossRef]
- Kamberov, E.; Sleptsova, I.; Suchyta, S.; Bruening, E.; Zeihler, W.; Nagel, J.S.; Langmore, J.; Makarov, V. Use of in vitro OmniPlex Libraries for high-throughput comparative genomics and molecular haplotyping. Proc. SPIE 2002, 4626, 340–351. [Google Scholar]
- Langmore, J.P. Rubicon Genomics, Inc. Pharmacogenomics 2002, 3, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Lizardi, P.M.; Huang, X.; Zhu, Z.; Bray-Ward, P.; Thomas, D.C.; Ward, D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998, 19, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Dean, F.B.; Hosono, S.; Fang, L.; Wu, X.; Faruqi, A.F.; Bray-Ward, P.; Sun, Z.; Zong, Q.; Du, Y.; Du, J.; et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 2002, 99, 5261–5266. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.L.; Hansen, M.S.; Faruqi, A.F.; Giannola, D.; Irsula, O.R.; Lasken, R.S.; Latterich, M.; Makarov, V.; Oliphant, A.; Pinter, J.H.; et al. Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel. Genome Res. 2004, 14, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Bergen, A.W.; Haque, K.A.; Qi, Y.; Beerman, M.B.; Garcia-Closas, M.; Rothman, N.; Chanock, S.J. Comparison of yield and genotyping performance of multiple displacement amplification and OmniPlex whole genome amplified DNA generated from multiple DNA sources. Hum. Mutat. 2005, 26, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Beaty, T.H.; Boyce, P.; Scott, A.F.; McIntosh, I. Comparing whole-genome amplification methods and sources of biological samples for single-nucleotide polymorphism genotyping. Clin. Chem. 2005, 51, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Pask, R.; Rance, H.E.; Barratt, B.J.; Nutland, S.; Smyth, D.J.; Sebastian, M.; Twells, R.C.; Smith, A.; Lam, A.C.; Smink, L.J.; et al. Investigating the utility of combining Phi29 whole genome amplification and highly multiplexed single nucleotide polymorphism BeadArray genotyping. BMC Biotechnol. 2004, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinard, R.; de Winter, A.; Sarkis, G.J.; Gerstein, M.B.; Tartaro, K.R.; Plant, R.N.; Egholm, M.; Rothberg, J.M.; Leamon, J.H. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genom. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.; Singh, A.K. Single cell genome sequencing. Curr. Opin. Biotechnol. 2011, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bergen, A.W.; Qi, Y.; Haque, K.A.; Welch, R.A.; Chanock, S.J. Effects of DNA mass on multiple displacement whole genome amplification and genotyping performance. BMC Biotechnol. 2005, 5. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.M.; Wiemels, J.L.; Wrensch, M.; Wiencke, J.K. DNA quantification of whole genome amplified samples for genotyping on a multiplexed bead array platform. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1686–1690. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Lin, M.; Beroukhim, R.; Lee, J.C.; Zhao, X.; Richter, D.J.; Gabriel, S.; Herman, P.; Sasaki, H.; Altshuler, D.; et al. Genome coverage and sequence fidelity of Phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004, 32. [Google Scholar] [CrossRef] [PubMed]
- Hosono, S.; Faruqi, A.F.; Dean, F.B.; Du, Y.; Sun, Z.; Wu, X.; Du, J.; Kingsmore, S.F.; Egholm, M.; Lasken, R.S. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003, 13, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.H.; Zhang, S.L.; Li, J.L.; Yap, W.S.; Howe, T.C.; Tan, B.P.; Lee, Y.S.; Wong, D.; Khoo, K.L.; Seto, K.Y.; et al. Using whole genome amplification (WGA) of low-volume biopsies to assess the prognostic role of EGFR, KRAS, p53, and CMET mutations in advanced-stage non-small cell lung cancer (NSCLC). J. Thorac. Oncol. 2009, 4, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Van Beers, E.H.; Joosse, S.A.; Ligtenberg, M.J.; Fles, R.; Hogervorst, F.B.; Verhoef, S.; Nederlof, P.M. A multiplex PCR predictor for aCGH success of FFPE samples. Br. J. Cancer 2006, 94, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- PolyPhen-2 prediction of functional effects of human nsSNPs. Available online: http://genetics.bwh.harvard.edu/pph2/ (accessed on 28 February 2015).
- SIFT Blink. Available online: http://sift.jcvi.org/ (accessed on 28 February 2015).
- 1000Genomes. Available online: http://www.1000genomes.org/ (accessed on 28 February 2015).
- Coco, S.; Truini, A.; Alama, A.; dal-Bello, M.G.; Venè, R.; Garuti, A.; Carminati, E.; Rijavec, E.; Genova, C.; Barletta, G.; et al. Afatinib resistance in non-small cell lung cancer involves the PI3K/AKT and MAPK/ERK signalling pathways and epithelial-to-mesenchymal transition. Target Oncol. 2015, 10, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Rehm, H.L.; Bale, S.J.; Bayrak-Toydemir, P.; Berg, J.S.; Brown, K.K.; Deignan, J.L.; Friez, J.; Funke, B.H.; Hegde, M.R.; Lyon, E.; et al. ACMG clinical laboratory standards for next-generation sequencing. Genet. Med. 2013, 15, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Coco, S.; de Mariano, M.; Valdora, F.; Servidei, T.; Ridola, V.; Andolfo, I.; Oberthuer, A.; Tonini, G.P.; Longo, L. Identification of ALK germline mutation (3605delG) in pediatric anaplastic medulloblastoma. J. Hum. Genet. 2012, 57, 682–684. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanni, I.; Coco, S.; Truini, A.; Rusmini, M.; Dal Bello, M.G.; Alama, A.; Banelli, B.; Mora, M.; Rijavec, E.; Barletta, G.; et al. Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGM™ Platform. Int. J. Mol. Sci. 2015, 16, 28765-28782. https://doi.org/10.3390/ijms161226129
Vanni I, Coco S, Truini A, Rusmini M, Dal Bello MG, Alama A, Banelli B, Mora M, Rijavec E, Barletta G, et al. Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGM™ Platform. International Journal of Molecular Sciences. 2015; 16(12):28765-28782. https://doi.org/10.3390/ijms161226129
Chicago/Turabian StyleVanni, Irene, Simona Coco, Anna Truini, Marta Rusmini, Maria Giovanna Dal Bello, Angela Alama, Barbara Banelli, Marco Mora, Erika Rijavec, Giulia Barletta, and et al. 2015. "Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGM™ Platform" International Journal of Molecular Sciences 16, no. 12: 28765-28782. https://doi.org/10.3390/ijms161226129
APA StyleVanni, I., Coco, S., Truini, A., Rusmini, M., Dal Bello, M. G., Alama, A., Banelli, B., Mora, M., Rijavec, E., Barletta, G., Genova, C., Biello, F., Maggioni, C., & Grossi, F. (2015). Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGM™ Platform. International Journal of Molecular Sciences, 16(12), 28765-28782. https://doi.org/10.3390/ijms161226129