tRNA Biology in Mitochondria
Abstract
:1. Introduction
2. The Pool of tRNAs in Mitochondria Consists of Encoded and Imported tRNAs
2.1. Distribution and Origin of tRNAs Encoded in Mitochondria
| Species | tRNA Genes Encoded by the mt-Genome | tRNA Genes Missing/not Expressed | Import Demonstrated | References | |||
|---|---|---|---|---|---|---|---|
| tRNA Content | tRNA Import | ||||||
| Metazoans | |||||||
| Mammals | |||||||
| Homo sapiens | 22 | 0 | Q | NC_012920 | [23] | ||
| Didelphis virginiana | 21 | K | K | NC_001610 | [24] | ||
| Molluscs | |||||||
| Crassostrea gigas | 18 | A, F, S | AF177226 | ||||
| Cnidarians | |||||||
| Metridium senile | 2 | all but 2 | NC_000933 | ||||
| Fungi | |||||||
| Ascomycotes | |||||||
| Saccharomyces cerevisiae | 24 | 0 | L, Q | NC_000933 | [25,26] | ||
| Chytridiomycotes | |||||||
| Spizellomyces punctatus | 8 | all but 8 | NC_003052 NC_003061 NC_003060 | ||||
| Plants | |||||||
| Bryophytes | |||||||
| Marchantia polymorpha | 29 | 0 | I(IAU) , T(AGU), V(AAC) | NC_001660 [27] | [28,29,30] | ||
| Angiosperms | |||||||
| Arabidopsis thaliana | 22 | all but 15 | F, W | NC_001284 | [31,32] | ||
| Nicotiana tabacum | 23 | at least 6 | A, G, V | NC_006581 | [33,34,35,36] | ||
| Solanum tuberosum | 20 | at least 7 | A, G, I, L, R, T, V | [37] | [38,39,40] | ||
| Triticum aestivum | 16 | 14 | A, G, H, I(IAU), L, R, V | NC_007579 | [41] | ||
| Gymnosperms | |||||||
| Larix leptoeuropaea | n.d | at least 11 | A, G, F, I(IAU), L, K, P, S(GCU), S(UGA), T, V | [39] | |||
| Chlorophytes | |||||||
| Chlamydomonas reinhardtii | 3 | all but 3 | for 31 tRNAs | NC_001638 | [42] | ||
| Scenedesmus obliquus | 27 | T | NC_002254 | ||||
| Nephroselmis olivacea | 26 | 0 | NC_008239 | ||||
| Other eukaryotes | |||||||
| Jakobides | |||||||
| Reclinomonas americana | 26 | T | NC_001823 | ||||
| Ciliophores | |||||||
| Tetrahymena pyriformis | 10 | all but 10 | for 26 tRNAs | NC_000862 | [43,44,45] | ||
| Trypanosomatides | |||||||
| Leishmania tarentolae | 0 | all | all except Q(CUG) | [46,47,48] | |||
| Trypanosoma brucei | 0 | all | all except initiator M and U | [22,49,50] | |||
| Apicomplexans | |||||||
| Plasmodium falciparum | 0 | all | C, F | NC_002375 | [51] | ||
2.2. Mitochondrial Import of tRNAs
2.2.1. Determinants for tRNA Import
2.2.2. Compared Levels of Imported tRNAs
2.2.3. Mechanistic Insights into tRNA Import

3. Remarkable Structural Features of Mitochondrial tRNAs
3.1. Sequences and Secondary Structure
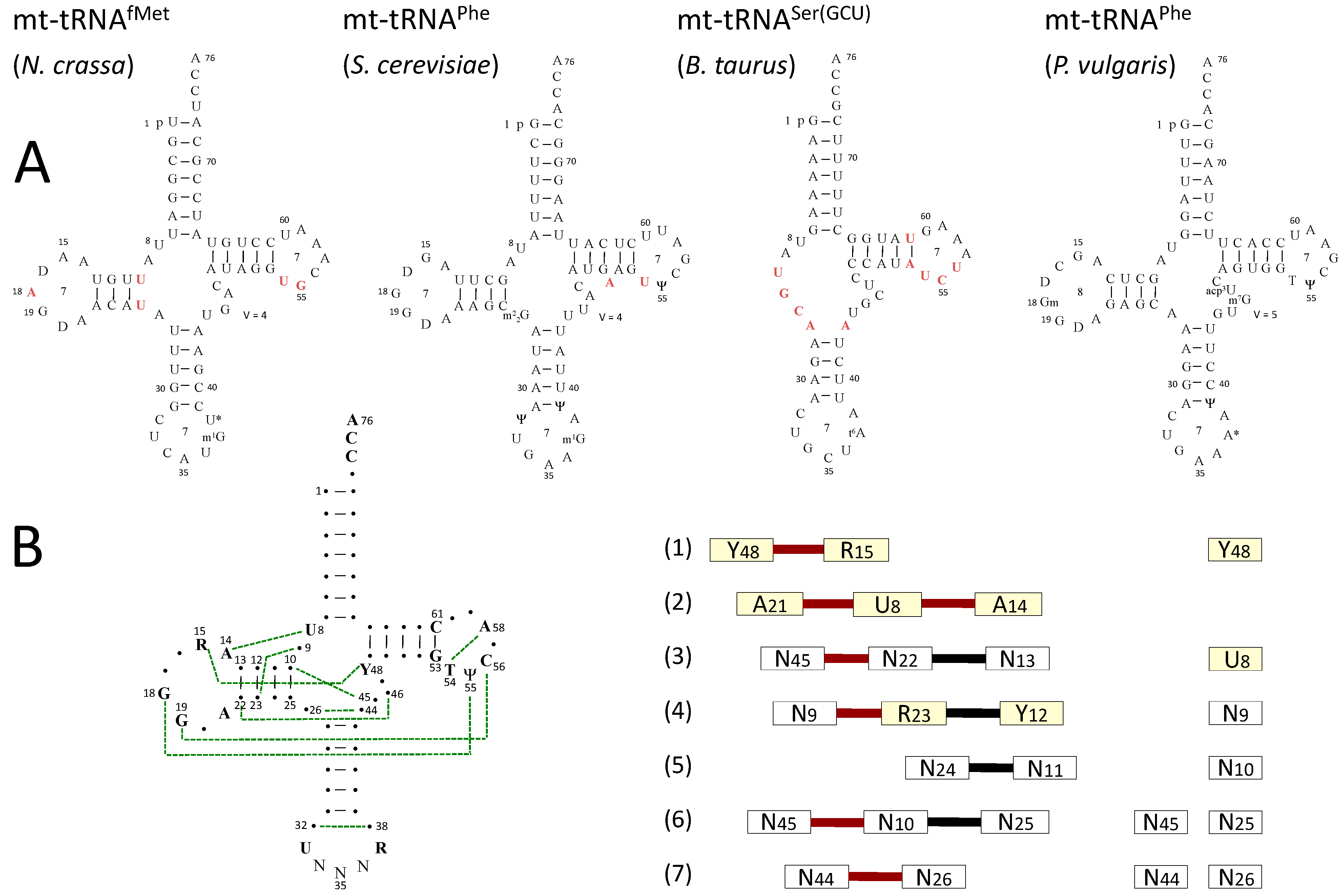
3.2. Post-Transcriptional Modifications in mt-tRNAs

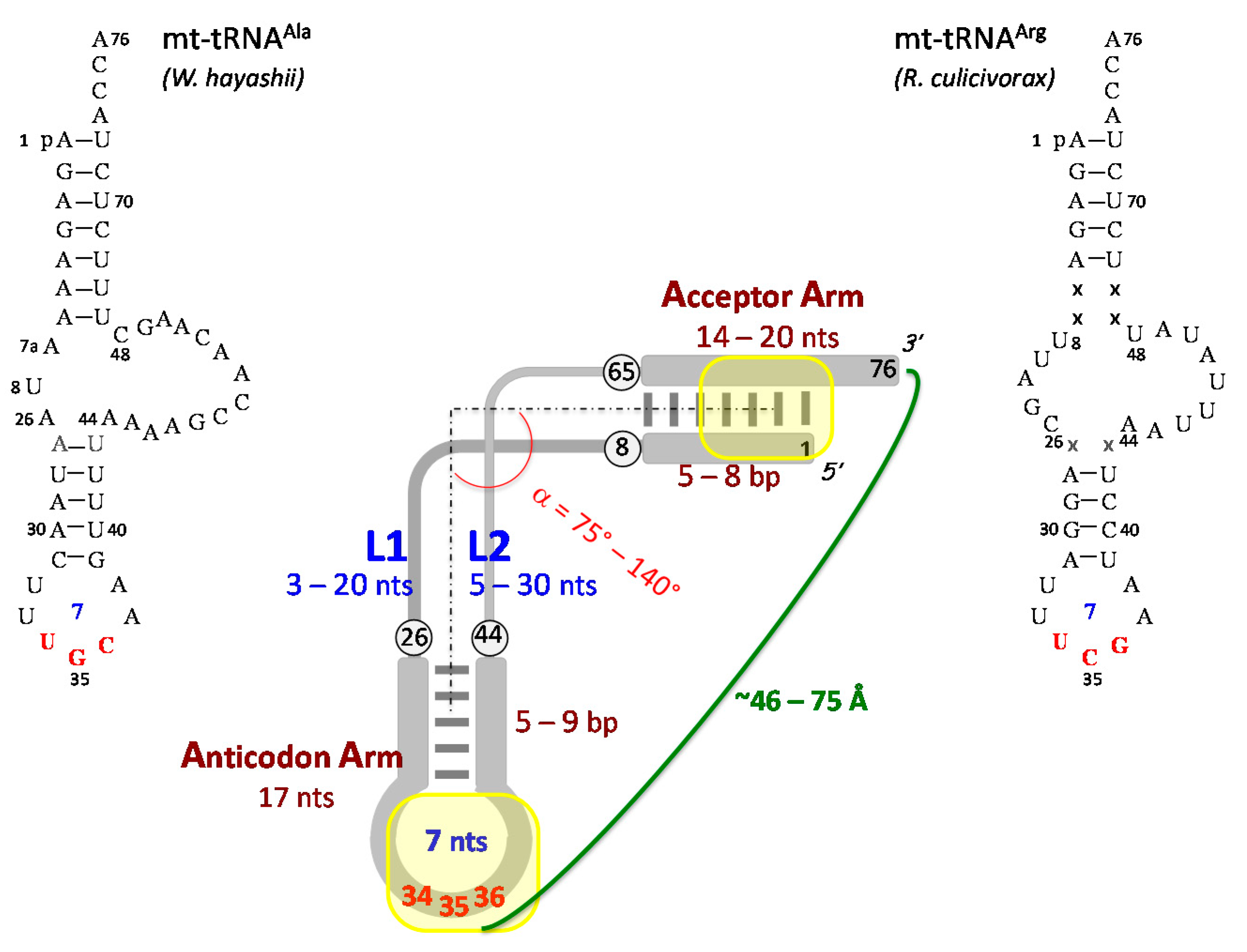
3.3. Three-Dimensional Structure of Free and Ligand-Bound mt-tRNAs
3.3.1. Free mt-tRNAs
| tRNA Groups | Group 1 | Group 2 | Group 3 | Group 4 | Group 5 |
|---|---|---|---|---|---|
| Eukaryote groups | Alveolates & Amoebas * Fungi Plants | Alveolates & Amoebas * some Fungi & Plants Metazoans | Metazoans (e.g., nematodes, arachnids & some bryozoan species) | Metazoans (some insect & bryozoan species & mammals) | Metazoans (acaria & some nematodes) |
| Representative tRNAs | Ath mt-Asp Sce mt-Glu Pca mt-LeuUAA Pvu mt-Phe | Mpo mt-Ala Hsa mt-Asp Pca mt-Cys Hsa mt-Phe | Asu mt-Ala Asu mt-Asn Cel mt-Asp Asu mt-Thr | Dno mt-Cys Bta mt-SerGCU Dya mt-SerGCU Pvi mt-SerGCU | Wha mt-Ala Rcu mt-Arg Rcu mt-Ile Rcu mt-Thr |
| Acceptor arm | canonical (stem: 7 bp) | canonical (stem: 7 bp) | quasi-canonical (stem: 4–7 bp) | canonical (stem: 7 bp) | atypical (stem: 4–5 bp) |
| Anticodon arm | canonical (stem: 5 bp & loop: 7 nts) | canonical (stem: 5 bp & loop: 7 nts) | canonical (stem: 5 bp & loop: 7 nts) | canonical (stem: 5 bp & loop: 7 nts) | atypical (stem: 5–9 bp) |
| D- and T-arms | canonical ** | atypical *** | atypical T-armless (atypical D-loops of 5–7 nts) | Atypical D-armless (atypical T-loops of 6–7 nts) | – D-/T-armless |
| T-arm | stem: 5 bp loop: 7 nts | stem: 4–5 bp loop: 6–7 nts | – | stem: 4–5 bp | – |
| D-arm | stem: 4 bp loop: 8–9 nts | stem: 4 bp **** | stem: 4 bp | – | – |
| L1 connector (size & characteristics) | 19–20 nts (with D-arm) | 16–20 nts (with D-arm) | 16–18 nts (with D-arm) | 5–12 nts | 3–11 nts |
| L2 connector (size & characteristics) | 21–30 nts (with VR & T-arm) | 19–22 nts (with VR & T-arm) | 6–7 nts | 20–21 nts | 5–14 nts |
| core organization in stacked base layers | 7 layers (canonical) | 7 layers (quasi-canonical) | 7 layers (atypical) | 6 layers (atypical) | 1 layer (atypical) |
3.3.2. mt-tRNAs Bound to aaRSs
3.3.3. mt-tRNAs Bound to Other Macromolecules and Macromolecular Assemblies
4. Biogenesis of Functional Mitochondrial tRNAs
4.1. Transcription
4.2. Maturation of tRNA 5'- and 3'-Termini
4.2.1. Maturation at 5'-Terminus
4.2.2. Maturation at 3'-Terminus

4.3. Functions and Mechanisms for tRNA Modifications and Editing
4.3.1. Mitochondrial tRNA Modifications
4.3.2. Mitochondrial tRNA Editing
5. Aminoacylation of Mitochondrial tRNAs
5.1. Global Features of Mitochondrial tRNA Aminoacylation Systems
5.2. Aminoacylation Identity of mt-tRNAs
5.2.1. General Considerations
5.2.2. Understanding mt-tRNA Identities for Aminoacylations Catalyzed by Class I aaRSs
5.2.3. Understanding mt-tRNA Identities for Aminoacylations Catalyzed by Class II aaRSs
5.2.4. Conserved Features and Peculiarities in mt-tRNA Aminoacylation Identity: An Overview
6. Conclusions and Perspectives
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Russell, O.; Turnbull, D. Mitochondrial DNA disease-molecular insights and potential routes to a cure. Exp. Cell Res. 2014, 325, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Schimper, A.F.W. Über die Entwicklung der Chlorophyllkörner und Farbkörper. Bot. Zeitung 1883, 41, 105–114, 121–131, 137–146, 153–162. [Google Scholar]
- Margulis, L. Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp. Soc. Exp. Biol. 1975, 29, 21–38. [Google Scholar] [PubMed]
- Gray, M.W.; Burger, G.; Lang, B.F. Mitochondrial evolution. Science 1999, 283, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. The pre-endosymbiont hypothesis: A new perspective on the origin and evolution of mitochondria. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Adams, K.L.; Palmer, J.D. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef] [PubMed]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.; Rehling, P.; van der Laan, M. Mitochondrial protein import: Common principles and physiological networks. Biochim. Biophys. Acta 2013, 1833, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; Duchêne, A.-M.; Maréchal-Drouard, L. Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 2008, 33, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, J.D.; Söll, D. Mitochondrial tRNA import—The challenge to understand has just begun. Biol. Chem. 2009, 390, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.A.; Hopper, A.K. Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip. Rev. RNA. 2011, 2, 802–817. [Google Scholar] [CrossRef] [PubMed]
- Bestwick, M.L.; Shadel, G.S. Accessorizing the human mitochondrial transcription machinery. Trends Biochem. Sci. 2013, 38, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Hammani, K.; Giegé, P. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014, 19, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Huot, J.L.; Enkler, L.; Megel, C.; Karim, L.; Laporte, D.; Becker, H.D.; Duchêne, A.-M.; Sissler, M.; Maréchal-Drouard, L. Idiosyncrasies in decoding mitochondrial genomes. Biochimie 2014, 100, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Maréchal-Drouard, L. Mitochondrial tRNA import: Are there distinct mechanisms? Trends Cell Biol. 2000, 10, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Bullerwell, C.E.; Gray, M.W. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete fungus. J. Biol. Chem. 2005, 280, 2463–2470. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; El Farouk-Ameqrane, S.; Ubrig, E.; Sauter, C.; Duchêne, A.-M.; Maréchal-Drouard, L. Molecular basis for the differential interaction of plant mitochondrial VDAC proteins with tRNAs. Nucleic Acids Res. 2014, 42, 9937–9948. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef] [PubMed]
- Fey, J.; Dietrich, A.; Cosset, A.; Desprez, T.; Maréchal-Drouard, L. Evolutionary aspects of “Chloroplast-like” trnN and trnH expression in higher-plant mitochondria. Curr. Genet. 1997, 32, 358–360. [Google Scholar] [CrossRef] [PubMed]
- Kitazaki, K.; Kubo, T.; Kagami, H.; Matsumoto, T.; Fujita, A.; Matsuhira, H.; Matsunaga, M.; Mikami, T. A horizontally transferred tRNACys gene in the sugar beet mitochondrial genome: Evidence that the gene is present in diverse angiosperms and its transcript is aminoacylated. Plant J. 2011, 68, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 2011, 80, 1033–1053. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.A.; Rinehart, J.J.; Krett, B.; Duvezin-Caubet, S.; Reichert, A.S.; Söll, D.; Alfonzo, J.D. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Nalt. Acad. Sci. USA 2008, 105, 9186–9191. [Google Scholar] [CrossRef]
- Dörner, M.; Altmann, M.; Pääbo, S.; Mörl, M. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol. Biol. Cell 2001, 12, 2688–2698. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.P.; Schneller, J.-M.; Stahl, A.J.; Dirheimer, G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry 1979, 18, 4600–4605. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, J.; Krett, B.; Rubio, M.-A.T.; Alfonzo, J.D.; Söll, D. Saccharomyces cerevisiae imports the cytosolic pathway for gln-tRNA synthesis into the mitochondion. Genes Dev. 2005, 19, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Yamato, K.; Ohta, E.; Nakamura, Y.; Takemura, M.; Nozato, N.; Akashi, K.; Ohyama, K. Transfer RNA genes in the mitochondrial genome from a liverwort, Marchantia polymorpha: The absence of chloroplast-like tRNAs. Nucleic Acids Res. 1992, 20, 3773–3777. [Google Scholar] [CrossRef] [PubMed]
- Akashi, K.; Sakurai, K.; Hirayama, J.; Fukuzawa, H.; Ohyama, K. Occurrence of nuclear-encoded tRNAIle in mitochondria of the liverwort Marchantia polymorpha. Curr. Genet. 1996, 30, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Akashi, K.; Hirayama, J.; Takenaka, M.; Yamaoka, S.; Suyama, Y.; Fukuzawa, H.; Ohyama, K. Accumulation of nuclear-encoded tRNAThr(AGU) in mitochondria of the liverwort Marchantia polymorpha. Biochim. Biophys. Acta 1997, 1350, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Akashi, K.; Takenaka, M.; Yamaoka, S.; Suyama, Y.; Fukuzawa, H.; Ohyama, K. Coexistence of nuclear DNA-encoded tRNAVal(AAC) and mitochondrial DNA-encoded tRNAVal(UAC) in mitochondria of a liverwort Marchantia polymorpha. Nucleic Acids Res. 1998, 26, 2168–2172. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Viry-Moussaid, M.; Dietrich, A.; Wintz, H. Evolution of a mitochondrial tRNAPhe gene in A. thaliana: Import of cytosolic tRNA phe into mitochondria. Biochem. Biophys. Res. Commun. 1997, 237, 432–437. [Google Scholar] [CrossRef]
- Duchêne, A.-M.; Maréchal-Drouard, L. The chloroplast-derived trnw and trnM-e genes are not expressed in Arabidopsis mitochondria. Biochem. Biophys. Res. Commun. 2001, 285, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, A.; Maréchal-Drouard, L.; Carneiro, V.; Cosset, A.; Small, I. A single base change prevents import of cytosolic tRNAAla into mitochondria in transgenic plants. Plant J. 1996, 10, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Delage, L.; Dietrich, A.; Cosset, A.; Maréchal-Drouard, L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell. Biol. 2003, 23, 4000–4012. [Google Scholar] [CrossRef] [PubMed]
- Laforest, M.J.; Delage, L.; Maréchal-Drouard, L. The T-domain of cytosolic tRNAVal, an essential determinant for mitochondrial import. FEBS Lett. 2005, 579, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; Schaeffer, C.; Maréchal-Drouard, L.; Duchêne, A.-M. Sequence dependence of tRNAGly import into tobacco mitochondria. Biochimie 2005, 87, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Maréchal-Drouard, L.; Guillemaut, P.; Cosset, A.; Arbogast, M.; Weber, F.; Weil, J.-H.; Dietrich, A. Transfer RNAs of potato (Solanum tuberosum) mitochondria have different genetic origins. Nucleic Acids Res. 1990, 18, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Small, I.; Maréchal-Drouard, L.; Masson, J.; Pelletier, G.; Cosset, A.; Weil, J.-H.; Dietrich, A. In vivo import of a normal or mutagenized heterologous transfer RNA into mitochondria of transgenic plants: Towards novel ways of influencing mitochondrial gene expression? EMBO J. 1992, 11, 1291–1296. [Google Scholar] [PubMed]
- Kumar, R.; Maréchal-Drouard, L.; Akama, K.; Small, I. Striking differences in mitochondrial tRNA import between different plant species. Mol. Gen. Genet. 1996, 252, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Brubacher-Kauffmann, S.; Maréchal-Drouard, L.; Cosset, A.; Dietrich, A.; Duchêne, A.-M. Differential import of nuclear-encoded tRNAGly isoacceptors into Solanum tuberosum mitochondria. Nucleic Acids Res. 1999, 27, 2037–2042. [Google Scholar] [CrossRef] [PubMed]
- Glover, K.E.; Spencer, D.F.; Gray, M.W. Identification and structural characterization of nucleus-encoded transfer RNAs imported into wheat mitochondria. J. Biol. Chem. 2001, 276, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Vinogradova, E.; Salinas, T.; Cognat, V.; Remacle, C.; Maréchal-Drouard, L. Steady-state levels of imported tRNAs in Chlamydomonas mitochondria are correlated with both cytosolic and mitochondrial codon usages. Nucleic Acids Res. 2009, 37, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Suyama, Y. The origins of mitochondrial ribonucleic acids in Tetrahymena pyriformis. Biochemistry 1967, 6, 2829–2839. [Google Scholar] [CrossRef] [PubMed]
- Chiu, N.; Chiu, A.; Suyama, Y. Native and imported transfer RNA in mitochondria. J. Mol. Biol. 1975, 99, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Suyama, Y. Two dimensional polyacrylamide gel electrophoresis analysis of Tetrahymena mitochondrial tRNA. Curr. Genet. 1986, 10, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.M.; Suyama, Y.; Dewes, H.; Campbell, D.A.; Simpson, L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989, 17, 5427–5445. [Google Scholar] [CrossRef] [PubMed]
- Lye, L.F.; Chen, D.H.; Suyama, Y. Selective import of nuclear-encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol. Biochem. Parasitol. 1993, 58, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Chen, D.H.; Suyama, Y. A nuclear tRNA gene cluster in the protozoan Leishmania tarentolae and differential distribution of nuclear-encoded tRNAs between the cytosol and mitochondria. Mol. Biochem. Parasitol. 1994, 65, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Hancock, K.; Hajduk, S.L. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 1990, 265, 19208–19215. [Google Scholar] [PubMed]
- Schneider, A.; McNally, K.P.; Agabian, N. Nuclear-encoded mitochondrial tRNAs of Trypanosoma brucei have a modified cytidine in the anticodon loop. Nucleic Acids Res. 1994, 22, 3699–3705. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, A. Plasmodium falciparum mitochondria import tRNAs along with an active phenylalanyl-tRNA synthetase. Biochem. J. 2015, 465, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Entelis, N.; Kolesnikova, O.; Kazakova, H.; Brandina, I.; Kamenski, P.; Martin, R.P.; Tarassov, I. Import of nuclear encoded rnas into yeast and human mitochondria: Experimental approaches and possible biomedical applications. Genet. Eng. 2002, 24, 191–213. [Google Scholar]
- Duchêne, A.-M.; Pujol, C.; Maréchal-Drouard, L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr. Genet. 2009, 55, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lithgow, T.; Schneider, A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 799–817. [Google Scholar] [CrossRef] [PubMed]
- Sieber, F.; Duchêne, A.-M.; Maréchal-Drouard, L. Mitochondrial rna import: From diversity of natural mechanisms to potential applications. Int. Rev. Cell Mol. Biol. 2011, 287, 145–190. [Google Scholar] [PubMed]
- Kamenski, P.; Kolesnikova, O.; Jubenot, V.; Entelis, N.; Krasheninnikov, I.A.; Martin, R.P.; Tarassov, I. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol. Cell 2007, 26, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Entelis, N.; Brandina, I.; Kamenski, P.; Krasheninnikov, I.A.; Martin, R.P.; Tarassov, I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006, 20, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Kamenski, P.; Smirnova, E.; Kolesnikova, O.; Krasheninnikov, I.A.; Martin, R.P.; Entelis, N.; Tarassov, I. tRNA mitochondrial import in yeast: Mapping of the import determinants in the carrier protein, the precursor of mitochondrial lysyl-tRNA synthetase. Mitochondrion 2010, 10, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, C.P.; Cech, T.R. The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes Dev. 1996, 10, 2870–2880. [Google Scholar] [CrossRef] [PubMed]
- Crausaz Esseiva, A.; Maréchal-Drouard, L.; Cosset, A.; Schneider, A. The T-stem determines the cytosolic or mitochondrial localization of trypanosomal tRNAsMet. Mol. Biol. Cell 2004, 15, 2750–2757. [Google Scholar] [CrossRef] [PubMed]
- Bouzaidi-Tiali, N.; Aeby, E.; Charriere, F.; Pusnik, M.; Schneider, A. Elongation factor 1a mediates the specificity of mitochondrial tRNA import in T. brucei. EMBO J. 2007, 26, 4302–4312. [Google Scholar] [CrossRef]
- Delage, L.; Duchêne, A.-M.; Zaepfel, M.; Maréchal-Drouard, L. The anticodon and the D-domain sequences are essential determinants for plant cytosolic tRNAVal import into mitochondria. Plant J. 2003, 34, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Salinas, K.; Wierzbicki, S.; Zhou, L.; Schmitt, M.E. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 2005, 280, 11352–11360. [Google Scholar] [CrossRef] [PubMed]
- Kapushoc, S.T.; Alfonzo, J.D.; Simpson, L. Differential localization of nuclear-encoded tRNAs between the cytosol and mitochondrion in Leishmania tarentolae. RNA 2002, 8, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Suyama, Y.; Wong, S.; Campbell, D.A. Regulated tRNA import in Leishmania mitochondria. Biochim. Biophys. Acta 1998, 1396, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.H.; Pach, R.; Crausaz, A.; Ivens, A.; Schneider, A. tRNAs in Trypanosoma brucei: Genomic organization, expression, and mitochondrial import. Mol. Cell. Biol. 2002, 22, 3707–3717. [Google Scholar] [CrossRef] [PubMed]
- Entelis, N.S.; Kolesnikova, O.A.; Dogan, S.; Martin, R.P.; Tarassov, I.A. 5S rRNA and tRNA import into human mitochondria: Comparison of in vitro requirements. J. Biol. Chem. 2001, 276, 45642–45653. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; Duby, F.; Larosa, V.; Coosemans, N.; Bonnefoy, N.; Motte, P.; Maréchal-Drouard, L.; Remacle, C. Co-evolution of mitochondrial tRNA import and codon usage determines translational efficiency in the green alga Chlamydomonas. PLoS Genet. 2012, 8, e1002946. [Google Scholar] [CrossRef] [PubMed]
- Tarassov, I.; Entelis, N.; Martin, R.P. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J. Mol. Biol. 1995, 245, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikova, O.; Kazakova, H.; Comte, C.; Steinberg, S.; Kamenski, P.; Martin, R.P.; Tarassov, I.; Entelis, N. Selection of RNA aptamers imported into yeast and human mitochondria. RNA 2010, 16, 926–941. [Google Scholar] [CrossRef] [PubMed]
- Brandina, I.; Smirnov, A.; Kolesnikova, O.; Entelis, N.; Krasheninnikov, I.A.; Martin, R.P.; Tarassov, I. tRNA import into yeast mitochondria is regulated by the ubiquitin-proteasome system. FEBS Lett. 2007, 581, 4248–4254. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; Duchêne, A.-M.; Delage, L.; Nilsson, S.; Glaser, E.; Zaepfel, M.; Maréchal-Drouard, L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 18362–18367. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, A.; Small, I.; Cosset, A.; Weil, J.-H.; Maréchal-Drouard, L. Editing and import: Strategies for providing plant mitochondria with a complete set of functional transfer RNAs. Biochimie 1996, 78, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Sieber, F.; Placido, A.; El Farouk-Ameqrane, S.; Duchêne, A.-M.; Maréchal-Drouard, L. A protein shuttle system to target RNA into mitochondria. Nucleic Acids Res. 2011, 39, e96. [Google Scholar] [CrossRef] [PubMed]
- Schekman, R. Editorial expression of concern and correction. Proc. Nalt. Acad. Sci. USA 2010, 107, 9476. [Google Scholar] [CrossRef]
- Pusnik, M.; Charriere, F.; Maser, P.; Waller, R.F.; Dagley, M.J.; Lithgow, T.; Schneider, A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol. Biol. Evol. 2009, 26, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Seidman, D.; Johnson, D.; Gerbasi, V.; Golden, D.; Orlando, R.; Hajduk, S. Mitochondrial membrane complex that contains proteins necessary for tRNA import in Trypanosoma brucei. J. Biol. Chem. 2012, 287, 8892–8903. [Google Scholar] [CrossRef] [PubMed]
- Tschopp, F.; Charriere, F.; Schneider, A. In vivo study in trypanosoma brucei links mitochondrial transfer rna import to mitochondrial protein import. EMBO Rep. 2011, 12, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Heckman, J.E.; Hecker, L.I.; Schwartzbach, S.D.; Barnett, W.E.; Baumstark, B.; RajBhandary, U.L. Structure and function of initiator methionine tRNA from the mitochondria of Neurospora crassa. Cell 1978, 13, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Heckman, J.E.; Alzner-Deweerd, B.; RajBhandary, U.L. Interesting and unusual features in the sequence of Neurospora crassa mitochondrial tyrosine transfer RNA. Proc. Natl. Acad. Sci. USA 1979, 76, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Sibler, A.P.; Martin, R.P.; Dirheimer, G. The nucleotide sequence of yeast mitochondrial histidine-tRNA. FEBS Lett. 1979, 107, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.P.; Sibler, A.P.; Schneller, J.-M.; Keith, G.; Stahl, A.J.; Dirheimer, G. Primary structure of yeast mitochondrial DNA-coded phenylalanine-tRNA. Nucleic Acids Res. 1978, 5, 4579–4592. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, L.; Guillemaut, P.; Grienenberger, J.-M.; Jeannin, G.; Weil, J.-H. Structure of bean mitochondrial transfer RNAPhe and localization of the transfer RNAPhe gene on the mitochondrial genomes of maize and wheat. FEBS Lett. 1985, 184, 289–293. [Google Scholar] [CrossRef]
- Maréchal, L.; Guillemaut, P.; Grienenberger, J.-M.; Jeannin, G.; Weil, J.-H. Sequence and codon recognition of bean mitochondria and chloroplast tRNAsTrp: Evidence for a high degree of homology. Nucleic Acid Res. 1985, 13, 4411–4416. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, L.; Guillemaut, P.; Weil, J.-H. Sequences of two bean mitochondria tRNAsTyr which differ in the level of post-transcriptional modification and have a prokaryotic-like large extra-loop. Plant Mol. Biol. 1985, 5, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Dirheimer, G.; Keith, G.; Sibler, A.-P.; Martin, R.P. The primary structure of tRNAs and their rare nucleosides. In Transfer RNA: Structure, Properties and Recognition; Schimmel, P., Söll, D., Abelson, J., Eds.; Cold Spring Harbor Lab.: Woodbury, NJ, USA, 1979; pp. 19–41. [Google Scholar]
- RajBhandary, U.L.; Heckman, J.E.; Yin, S.; Alzner-DeWeerd, B.; Ackerman, E. Recent develpments in tRNA sequencing methods as applied to analyses of mitochondrial tRNAs. In Transfer RNA: Structure, Properties and Recognition; Schimmel, P., Söll, D., Abelson, J., Eds.; Cold Spring Harbor Lab.: Woodbury, NJ, USA, 1979; pp. 3–17. [Google Scholar]
- Arcari, P.; Brownlee, G.G. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980, 8, 5207–5212. [Google Scholar] [CrossRef] [PubMed]
- De Bruijn, M.H.; Schreier, P.H.; Eperon, I.C.; Barrell, B.G.; Chen, E.Y.; Armstrong, P.W.; Wong, J.F.; Roe, B.A. A mammalian mitochondrial serine transfer RNA lacking the “Dihydrouridine” loop and stem. Nucleic Acids Res. 1980, 8, 5213–5222. [Google Scholar] [CrossRef] [PubMed]
- Pütz, J.; Dupuis, B.; Sissler, M.; Florentz, C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA 2007, 13, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA modification database, RNAmdb: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Jühling, F.; Pütz, J.; Stadler, P.; Sauter, C.; Florentz, C. Structure of transfer RNAs: Similarity and variability. Wiley Interdiscip. Rev. RNA 2012, 3, 37–61. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R.; Macfarlane, J.L.; Okimoto, R.; Clary, D.O.; Wahleithner, J.A. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc. Natl. Acad. Sci. USA 1987, 84, 1324–1328. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Pütz, J.; Florentz, C.; Stadler, P.F. Armless mitochondrial tRNAs in Enoplea (Nematoda). RNA Biol. 2012, 9, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Palopoli, M.F.; Minot, S.; Pei, D.; Satterly, A.; Endrizzi, J. Complete mitochondrial genomes of the human follicle mites Demodex brevis and D. folliculorum: Novel gene arrangement, truncated tRNA genes, and ancient divergence between species. BMC Genomics 2014, 15, 1124. [Google Scholar]
- Wende, S.; Platzer, E.G.; Juhling, F.; Pütz, J.; Florentz, C.; Stadler, P.F.; Mörl, M. Biological evidence for the world’s smallest tRNAs. Biochimie 2013, 100, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-S.; Jin, P.Y.; Zhang, K.J.; Ding, X.L.; Yang, S.X.; Ju, J.F.; Zhao, J.Y.; Hong, X.Y. The complete mitochondrial genomes of six species of Tetranychus provide insights into the phylogeny and evolution of spider mites. PLoS One 2014, 9, e110625. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K. Unique features of animal mitochondrial translation systems—The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc. Jpn. Acad. Series B-Phys. Biol. Sci. 2010, 86, 11–39. [Google Scholar] [CrossRef]
- Grosjean, H. DNA and RNA Modification Enzymes: Structure, Mechanisms, Function, and Evolution; Landes Bioscience: Austin, TX, USA, 2009; pp. 1–653. [Google Scholar]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. Modomics: A database of rna modification pathways–2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar]
- Suzuki, T.; Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014, 42, 7346–7357. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Moriya, J.; Yokogawa, T.; Wakita, K.; Ueda, T.; Nishikawa, K.; Crain, P.F.; Hashizume, T.; Pomerantz, S.C.; MacCloskey, J.A.; Kawai, G.; et al. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry 1994, 33, 2234–2239. [Google Scholar]
- Watanabe, Y.; Tsurui, H.; Ueda, T.; Furushima, R.; Takamiya, S.; Kita, K.; Nishikawa, K.; Watanabe, K. Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J. Biol. Chem. 1994, 269, 22902–22906. [Google Scholar] [PubMed]
- Suzuki, T.; Suzuki, T.; Wada, T.; Saigo, K.; Watanabe, K. Taurine as a constituent of mitochondrial tRNAs: New insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002, 21, 6581–6589. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Dietrich, A.; Weil, J.-H.; Maréchal-Drouard, L. A potato mitochondrial isoleucine tRNA is coded by a mitochondrial gene possessing a methionine anticodon. Nucleic Acid Res. 1990, 18, 5027–5030. [Google Scholar]
- Dubin, D.T.; HsuChen, C.-C.; Cleaves, G.R.; Timko, K.D. Sequence and structure of a serine transfer RNA with GCU anticodon from mosquito mitochondria. J. Mol. Biol. 1984, 176, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miyauchi, K.; Yokobori, S.I.; Shigi, N.; Kondow, A.; Takeuchi, N.; Yamagishi, A.; Watanabe, K. Taurine-containing uridine modifications in tRNA anticodons are required to decipher non-universal genetic codes in ascidian mitochondria. J. Biol. Chem. 2011, 286, 35494–35498. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Brulé, H.; Degoul, F.; Cepanec, C.; Leroux, J.-P.; Giegé, R.; Florentz, C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Voigts-Hoffmann, F.; Hengesbach, M.; Kobitski, A.Y.; van Aerschot, A.; Herdewijn, P.; Nienhaus, G.U.; Helm, M. A methyl group controls conformational equilibrium in human mitochondrial tRNALys. J. Am. Chem. Soc. 2007, 129, 13382–13383. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, I.; Kawai, G.; Watanabe, K. Higher-order structure and thermal instability of bovine mitochondrial tRNASer(UGA) investigated by proton NMR spectroscopy. J. Mol. Biol. 1998, 284, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, S.; Cedergren, R. Structural compensation in atypical mitochondrial tRNAs. Nat. Struct. Biol. 1994, 1, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, S.; Leclerc, F.; Cedergren, R. Structural rules and conformational compensations in the tRNA L-form. J. Mol. Biol. 1997, 266, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.F.; Lavrov, D.; Beck, N.; Steinberg, S.V. Mitochondrial tRNA structure, identity, and evolution of the genetic code. In Organelle Genetics. Evolution of Organelle Genomes and Gene Expression; Bullerwell, C.E., Ed.; Springer-Verlag: Berlin, Germany, 2012; pp. 431–474. [Google Scholar]
- Leehey, M.A.; Squassoni, C.A.; Friederich, M.W.; Mills, J.B.; Hagerman, P.J. A noncanonical tertiary conformation of a human mitochondrial transfer RNA. Biochemistry 1995, 34, 16235–16239. [Google Scholar] [CrossRef] [PubMed]
- Frazer-Abel, A.A.; Hagerman, P.J. Determination of the angle between the acceptor and anticodon stems of a truncated mitochondrial tRNA. J. Mol. Biol. 1999, 285, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, K.; O'Donoghue, P.; Gundllapalli, S.; Araiso, Y.; Ishitani, R.; Umehara, T.; Söll, D.; Nureki, O. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature 2009, 457, 1163–1167. [Google Scholar] [CrossRef] [PubMed]
- Théobald-Dietrich, A.; Frugier, M.; Giegé, R.; Rudinger-Thirion, J. Atypical archaeal tRNA pyrrolysine transcript behaves towards EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 2004, 32, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuki, T.; Kawai, G.; Watanabe, K. The minimal tRNA: Unique structure of Ascaris suum mitochondrial tRNASer(UCU) having a short T arm and lacking the entire D arm. FEBS Lett. 2002, 514, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Messmer, M.; Pütz, J.; Suzuki, T.; Suzuki, T.; Sauter, C.; Sissler, M.; Florentz, C. Tertiary network in mammalian mitochondrial tRNAAsp revealed by solution probing and phylogeny. Nucleic Acids Res. 2009, 37, 6881–6895. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, A.D.; Khvorova, A.M.; Sauter, C.; Florentz, C.; Giegé, R. Mimics of yeast tRNAAsp and their recognition by aspartyl-tRNA synthetase. Biochemistry 1999, 38, 11926–11932. [Google Scholar] [CrossRef] [PubMed]
- Lusic, H.; Gustilo, E.M.; Vendeix, F.A.; Kaiser, R.; Delaney, M.O.; Graham, W.D.; Moye, V.A.; Cantara, W.A.; Agris, P.F.; Deiters, A. Synthesis and investigation of the 5-formylcytidine modified, anticodon stem and loop of the human mitochondrial tRNAMet. Nucleic Acids Res. 2008, 36, 6548–6557. [Google Scholar] [CrossRef] [PubMed]
- Bilbille, Y.; Gustilo, E.M.; Harris, K.A.; Jones, C.N.; Lusic, H.; Kaiser, R.J.; Delaney, M.O.; Spremulli, L.L.; Deiters, A.; Agris, P.F. The human mitochondrial tRNAMet: Structure/function relationship of a unique modification in the decoding of unconventional codons. J. Mol. Biol. 2011, 406, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Murphy, F.V.; Demirci, H.; Agris, P.F. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl. Acad. Sci. USA 2013, 110, 10964–10969. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Mörl, M.; Hartmann, R.; Sprinzl, M.; Stadler, P.F.; Pütz, J. Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef] [PubMed]
- Schnare, M.N.; Heinonen, T.Y.; Young, P.G.; Gray, M.W. Phenylalanine and tyrosine transfer RNAs encoded by Tetrahymena pyriformis mitochondrial DNA: Primary sequence, post-transcriptional modifications, and gene localization. Curr. Genet. 1985, 9, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Cognat, V.; Pawlak, G.; Duchêne, A.-M.; Daujat, M.; Gigant, A.; Salinas, T.; Michaud, M.; Gutmann, B.; Giegé, P.; Gobert, A.; et al. PlantRNA, a database for tRNAs of photosynthetic eukaryotes. Nucleic Acids Res. 2013, 41, D273–D279. [Google Scholar]
- Schnare, M.N.; Greenwood, S.J.; Gray, M.W. Primary sequence and post-transcriptional modification pattern of an unusual mitochondrial tRNAMet from Tetrahymena pyriformis. FEBS Lett. 1995, 362, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Masta, S.E.; Boore, J.L. Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol. Biol. Evol. 2008, 25, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Masta, S.E.; McCall, A.; Longhorm, S.J. Rare genomic changes and mitochondrial sequences provide independent support for congruent relationships among the sea spiders (Arthropoda, Pycnogonida). 2010, 57, 59–70. [Google Scholar]
- Auffinger, P.; Westhof, E. An extended structural signature for the tRNA anticodon loop. RNA 2001, 7, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Durant, P.C.; Davis, D.R. Stabilization of the anticodon stem-loop of tRNALys, 3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999, 31, 115–131. [Google Scholar] [CrossRef]
- Klipcan, L.; Moor, N.; Finarov, I.; Kessler, N.; Sukhanova, M.; Safro, M.G. Crystal structure of human mitochondrial phers complexed with tRNAPhe in the active “Open” state. J. Mol. Biol. 2012, 415, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Chimnaronk, S.; Gravers Jeppesen, M.; Suzuki, T.; Nyborg, J.; Watanabe, K. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005, 24, 3369–3379. [Google Scholar] [CrossRef] [PubMed]
- Neuenfeldt, A.; Lorber, B.; Ennifar, E.; Gaudry, A.; Sauter, C.; Sissler, M.; Florentz, C. Thermodynamic properties distinguish human mitochondrial aspartyl-tRNA synthetase from bacterial homolog with same 3D architecture. Nucleic Acids Res. 2013, 41, 2698–2708. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Roy, H.; Qin, D.; Rubio, M.A.; Alfonzo, J.D.; Fredrick, K.; Ibba, M. Pathogenic mechanism of a human mitochondrial tRNAPhe mutation associated with myoclonic epilepsy with ragged red fibers syndrome. Proc. Natl. Acad. Sci. USA 2007, 104, 15299–15304. [Google Scholar] [CrossRef] [PubMed]
- Goldgur, Y.; Mosyak, L.; Reshetnikova, L.; Ankilova, V.; Lavrik, O.; Khodyreva, S.; Safro, M. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure 1997, 5, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Pinker, F.; Fuchsbauer, O.; Gutmann, B.; Boutin, R.; Roblin, P.; Sauter, C.; Giegé, P. Structural insights into protein-only RNase P complexed with tRNA. Nat. Commun. 2013, 4, 1353. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.K.; Sharma, M.R. Structural aspects of mitochondrial translational apparatus. Curr. Opin. Struct. Biol. 2012, 22, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Greber, B.J.; Boehringer, D.; Leitner, A.; Bieri, P.; Voigts-Hoffmann, F.; Erzberger, J.P.; Leibundsgut, M.; Aebersold, M.; Ban, N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 2014, 505, 515–519. [Google Scholar]
- Amunts, A.; Brown, A.; Bai, X.C.; Llacer, J.L.; Hussain, T.; Emsley, P.; Long, F.; Murshudov, G.; Scheres, S.H.; Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 2014, 343, 1485–1489. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; Bankier, A.T.; Barrel, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, J.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, W. Of P and Z: Mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta 2012, 1819, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Ringel, R.; Sologub, M.; Morozov, Y.I.; Litonin, D.; Cramer, P.; Temiakov, D. Structure of human mitochondrial RNA polymerase. Nature 2011, 478, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Schwinghammer, K.; Cheung, A.C.; Morozov, Y.I.; Agaronyan, K.; Temiakov, D.; Cramer, P. Structure of human mitochondrial RNA polymerase elongation complex. Nat. Struct. Mol. Biol. 2013, 20, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Giegé, P. Pentatricopeptide repeat proteins: A set of modular RNA-specific binders massively used for organelle gene expression. RNA Biol. 2013, 10, 1417–1418. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K.; Richter, U.; Meyer, E.H.; Delannoy, E.; de Longevialle, A.F.; O'Toole, N.; Borner, T.; Millar, A.H.; Small, I.D.; Whelan, J. Phage-type RNA polymerase rpotmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 2009, 21, 2762–2779. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Gobert, A.; Giegé, P. Mitochondrial genome evolution and the emergence of PPR proteins. In Mitochondrial Genome Evolution; Drouard, L., Ed.; Elsevier: Amsterdam, Netherlands, 2012; pp. 253–313. [Google Scholar]
- Lang, B.F.; Burger, G.; O’Kelly, C.J.; Cedergren, R.; Golding, G.B.; Lemieux, C.; Sankoff, D.; Turmel, M.; Gray, M.W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 1997, 387, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Altman, S. A view of RNase P. Mol. Biosyst. 2007, 3, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Pinker, F.; Bonnard, G.; Gobert, A.; Gutmann, B.; Hammani, K.; Sauter, C.; Gegenheimer, P.A.; Giegé, P. PPR proteins shed a new light on RNase P biology. RNA Biol. 2013, 10, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Seif, E.R.; Forget, L.; Martin, N.C.; Lang, B.F. Mitochondrial RNase P RNAs in ascomycete fungi: Lineage-specific variations in RNA secondary structure. RNA 2003, 9, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Willkomm, D.K.; Schön, A.; Hartmann, R.K. RNase P of the Cyanophora paradoxa cyanelle: A plastid ribozyme. Biochimie 2007, 89, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Burger, G.; Gray, M.W.; Forget, L.; Lang, B.F. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol. Evol. 2013, 5, 418–438. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, E.; Hartmann, R.K. The enigma of ribonuclease P evolution. Trends Genet. 2003, 19, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, J.; Frank, P.; Loffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Gutmann, B.; Taschner, A.; Gossringer, M.; Holzmann, J.; Hartmann, R.K.; Rossmanith, W.; Giegé, P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010, 17, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Taschner, A.; Weber, C.; Buzet, A.; Hartmann, R.K.; Hartig, A.; Rossmanith, W. Nuclear RNase P of Trypanosoma brucei: A single protein in place of the multicomponent RNA-protein complex. Cell Rep. 2012, 2, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Sugita, C.; Komura, Y.; Tanaka, K.; Kometani, K.; Satoh, H.; Sugita, M. Molecular characterization of three PRORP proteins in the moss Physcomitrella patens: Nuclear PRORP protein is not essential for moss viability. PLoS One 2014, 9, e108962. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Gobert, A.; Giegé, P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012, 26, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, L.V.; Gossringer, M.; Weber, C.; Buzet, A.; Rossmanith, W.; Hartmann, R.K. tRNA processing by protein-only versus RNA-based RNase P: Kinetic analysis reveals mechanistic differences. Chembiochem. 2012, 13, 2270–2276. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Schilling, O.; Spath, B.; Marchfelder, A. The tRNase Z family of proteins: Physiological functions, substrate specificity and structural properties. Biol. Chem. 2005, 386, 1253–1264. [Google Scholar] [PubMed]
- Rossmanith, W. Localization of human RNase Z isoforms: Dual nuclear/mitochondrial targeting of the elac2 gene product by alternative translation initiation. PLoS One 2011, 6, e19152. [Google Scholar] [CrossRef] [PubMed]
- Duchêne, A.-M.; Giegé, P. Dual localized mitochondrial and nuclear proteins as gene expression regulators in plants. Front. Plant Sci. 2012, 3, 221. [Google Scholar] [CrossRef] [PubMed]
- Canino, G.; Bocian, E.; Barbezier, N.; Echeverria, M.; Forner, J.; Binder, S.; Marchfelder, A. Arabidopsis encodes four tRNase Z enzymes. Plant Physiol. 2009, 150, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Vörtler, S.; Mörl, M. tRNA-nucleotidyltransferases: Highly unusual RNA polymerases with vital functions. FEBS Lett. 2010, 584, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Reichert, A.S.; Thurlow, D.L.; Mörl, M. A eubacterial origin for the human tRNA nucleotidyltransferase? Biol. Chem. 2001, 382, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Hammani, K.; Bonnard, G.; Bouchoucha, A.; Gobert, A.; Pinker, F.; Salinas, T.; Giegé, P. Helical repeats modular proteins are major players for organelle gene expression. Biochimie 2014, 100, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxii gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Knoop, V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. 2011, 68, 567–586. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Alfonzo, J.D. Do all modifications benefit all tRNAs? FEBS Lett. 2010, 584, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, T.; Suzuki, T.; Ishii, N.; Ueda, T.; Ohta, S.; Watanabe, K. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000, 467, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Yasukawa, T.; Marjavaara, S.K.; Jacobs, H.T.; Holt, I.J.; Watanabe, K.; Suzuki, T. Acquisition of the wobble modification in mitochondrial tRNALeu(CUN) bearing the G12300A mutation suppresses the MELAS molecular defect. Hum. Mol. Gen. 2006, 15, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nagao, A. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip. Rev. RNA 2011, 2, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Umeda, N.; Suzuki, T.; Yukawa, M.; Ohya, Y.; Shindo, H.; Watanabe, K. Mitochondria-specific rna-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005, 280, 1613–1624. [Google Scholar]
- Vilardo, E.; Nachbagauer, C.; Buzet, A.; Taschner, A.; Holzmann, J.; Rossmanith, W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase--extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012, 40, 11583–11593. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, S.I.; Pääbo, S. tRNA editing in metazoans. Nature 1995, 377, 490. [Google Scholar] [CrossRef] [PubMed]
- Börner, G.V.; Yokobori, S.-I.; Mörl, M.; Dörner, M.; Pääbo, S. RNA editing in metazoan mitochondria: Staying fit without sex. FEBS Lett. 1997, 409, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Reichert, A.; Rothbauer, U.; Mörl, M. Processing and editing of overlapping tRNAs in human mitochondria. J. Biol. Chem. 1998, 273, 31977–31984. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, K.M.; Gray, M.W. Editing of transfer rnas in Acanthamoeba castellanii mitochondria. Science 1993, 259, 812–816. [Google Scholar] [CrossRef] [PubMed]
- Laforest, M.J.; Bullerwell, C.E.; Forget, L.; Lang, B.F. Origin, evolution, and mechanism of 5' tRNA editing in chytridiomycete fungi. RNA 2004, 10, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Antes, T.; Costandy, H.; Mahendran, R.; Spottswood, M.; Miller, D. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol. Cell. Biol. 1998, 18, 7521–7527. [Google Scholar] [PubMed]
- Börner, G.V.; Mörl, M.; Janke, A.; Pääbo, S. RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 1996, 15, 5949–5957. [Google Scholar] [PubMed]
- Giegé, P.; Brennicke, A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 1999, 96, 15324–15329. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Hartel, B.; Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Fey, J.; Weil, J.-H.; Tomita, K.; Cosset, A.; Dietrich, A.; Small, I.; Maréchal-Drouard, L. Editing of plant mitochondrial transfer RNAs. Acta Biochim. Pol. 2001, 48, 383–389. [Google Scholar] [PubMed]
- Maréchal-Drouard, L.; Cosset, A.; Remacle, C.; Ramamonjisoa, D.; Dietrich, A. A single editing event is a prerequisite for efficient processing of potato mitochondrial phenylalanine tRNA. Mol. Cell. Biol. 1996, 16, 3504–3510. [Google Scholar] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, J.D.; Blanc, V.; Estevez, A.M.; Rubio, M.A.; Simpson, L. C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999, 18, 7056–7062. [Google Scholar] [CrossRef] [PubMed]
- Aldinger, C.A.; Leisinger, A.K.; Gaston, K.W.; Limbach, P.A.; Igloi, G.L. The absence of A-to-I editing in the anticodon of plant cytoplasmic tRNAArg(ACG) demands a relaxation of the wobble decoding rules. RNA Biol. 2012, 9, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Crain, P.F.; Alfonzo, J.D.; Rozenski, J.; Kapushoc, S.T.; McCloskey, J.A.; Simpson, L. Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. RNA 2002, 8, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Bruske, E.I.; Sendfeld, F.; Schneider, A. Thiolated tRNAs of Trypanosoma brucei are imported into mitochondria and dethiolated after import. J. Biol. Chem. 2009, 284, 36491–36499. [Google Scholar] [CrossRef] [PubMed]
- Brindefalk, B.; Viklund, J.; Larsson, D.; Thollesson, M.; Andersson, S.G. Origin and evolution of the mitochondrial aminoacyl-tRNA synthetases. Mol. Biol. Evol. 2007, 24, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, L.; Fender, A.; Rudinger-Thirion, J.; Giegé, R.; Florentz, C.; Sissler, M. Towards the full set of human mitochondrial aminoacyl-tRNA synthetases: Characterization of AspRS and TyrRS. Biochemistry 2005, 44, 4805–4816. [Google Scholar] [CrossRef] [PubMed]
- Haen, K.M.; Pett, W.; Lavrov, D.V. Parallel loss of nuclear-encoded mitochondrial aminoacyl-tRNA synthetases and mtDNA-encoded tRNAs in Cnidaria. Mol. Biol. Evol. 2010, 27, 2216–2219. [Google Scholar] [CrossRef] [PubMed]
- Sanni, A.; Walter, P.; Boulanger, Y.; Ebel, J.-P.; Fasiolo, F. Evolution of aminoacyl-tRNA synthetase quaternary structure and activity: Saccharomyces cerevisiae mitochondrial phenylalanyl-tRNA synthetase. Proc. Natl. Acad. Sci. USA 1991, 88, 8387–8391. [Google Scholar] [CrossRef] [PubMed]
- Klipcan, L.; Finarov, I.; Moor, N.; Safro, M.G. Structural aspects of phenylalanylation and quality control in three major forms of phenylalanyl-tRNA synthetase. J. Amino Acids 2010, 2010. org/10.4061/2010/983503. [Google Scholar] [CrossRef]
- Brandao, M.M.; Silva-Filho, M.C. Evolutionary history of Arabidopsis thaliana aminoacyl-tRNA synthetase dual-targeted proteins. Mol. Biol. Evol. 2011, 28, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Duchêne, A.-M.; Giritch, A.; Hoffmann, B.; Cognat, V.; Lancelin, D.; Peeters, N.M.; Zaepfel, M.; Maréchal-Drouard, L.; Small, I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 16484–16489. [Google Scholar] [CrossRef] [PubMed]
- Bullard, J.M.; Cai, Y.C.; Spremulli, L.L. Expression and characterization of the human mitochondrial leucyl-tRNA synthetase. Biochim. Biophys. Acta 2000, 1490, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Ibba, M.; Söll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000, 69, 617–650. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Yokoyama, S. Two enzymes bound to one tRNA assume alternative conformations for consecutive reactions. Nature 2010, 467, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Pujol, C.; Bailly, M.; Kern, D.; Maréchal-Drouard, L.; Becker, H.; Duchêne, A.-M. Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic gln-tRNAGln synthesis in plants. Proc. Natl. Acad. Sci. USA 2008, 105, 6481–6485. [Google Scholar] [CrossRef] [PubMed]
- Fréchin, M.; Senger, B.; Braye, M.; Kern, D.; Martin, R.P.; Becker, H.D. Yeast mitochondrial gln-tRNAGln is generated by a GATfab-mediated transamidation pathway involving ARC1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 2009, 23, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Araiso, Y.; Huot, J.L.; Sekiguchi, T.; Frechin, M.; Fischer, F.; Enkler, L.; Senger, B.; Ishitani, R.; Becker, H.D.; Nureki, O. Crystal structure of Saccharomyces cerevisiae mitochondrial GATfab reveals a novel subunit assembly in tRNA-dependent amidotransferases. Nucleic Acids Res. 2014, 42, 6052–6063. [Google Scholar] [CrossRef] [PubMed]
- Nagao, A.; Suzuki, T.; Katoh, T.; Sakaguchi, Y. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc. Natl. Acad. Sci. USA 2009, 106, 16209–16214. [Google Scholar] [CrossRef] [PubMed]
- Echevarria, L.; Clemente, P.; Hernandez-Sierra, R.; Gallardo, M.E.; Fernandez-Moreno, M.A.; Garesse, R. Glutamyl-tRNAGln amidotransferase is essential for mammalian mitochondrial translation in vivo. Biochem. J. 2014, 460, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Mailu, B.M.; Ramasamay, G.; Mudeppa, D.G.; Li, L.; Lindner, S.E.; Peterson, M.J.; DeRocher, A.E.; Kappe, S.H.; Rathod, P.K.; Gardner, M.J. A nondiscriminating glutamyl-tRNA synthetase in the plasmodium apicoplast: The first enzyme in an indirect aminoacylation pathway. J. Biol. Chem. 2013, 288, 32539–32552. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, Y.; Himeno, H.; Miura, K.-I.; Watanabe, K. Unilateral aminoacylation specificity between bovine mitochondria and eubacteria. J. Biochem. (Tokyo) 1991, 109, 421–427. [Google Scholar]
- Fender, A.; Sauter, C.; Messmer, M.; Pütz, J.; Giegé, R.; Florentz, C.; Sissler, M. Loss of a primordial identity element for a mammalian mitochondrial aminoacylation system. J. Biol. Chem. 2006, 281, 15980–15986. [Google Scholar] [CrossRef] [PubMed]
- Charriere, F.; O’Donoghue, P.; Helgadottir, S.; Maréchal-Drouard, L.; Cristodero, M.; Horn, E.K.; Söll, D.; Schneider, A. Dual targeting of a tRNAAsp requires two different aspartyl-tRNA synthetases in Trypanosoma brucei. J. Biol. Chem. 2009, 284, 16210–16217. [Google Scholar] [CrossRef] [PubMed]
- Fender, A.; Gaudry, A.; Jühling, F.; Sissler, M.; Florentz, C. Adaptation of aminoacylation identity rules to mammalian mitochondria. Biochimie 2012, 94, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Eriani, G. Transfer RNA recognition and aminoacylation by synthetases. In Encyclopedia of Life Sciences (ELS); John Wiley & Sons, Ltd.: Chichester, UK, 2014. [Google Scholar]
- Janke, A.; Pääbo, S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993, 21, 1523–1525. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Sohm, B.; Frugier, M.; Brulé, H.; Olszak, K.; Przykorska, A.; Florentz, C. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 2003, 328, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Sohm, B.; Sissler, M.; Park, H.; King, M.P.; Florentz, C. Recognition of human mitochondrial tRNALeu(UUR) by its cognate leucyl-tRNA synthetase. J. Mol. Biol. 2004, 339, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Nonaka, I.; Horai, S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 1990, 348, 651–653. [Google Scholar] [CrossRef] [PubMed]
- van den Ouweland, J.M.W.; Lemkes, H.H.P.J.; Ruitenbeek, W.; Sandkuijl, L.A.; de Vijlder, M.F. Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1992, 1, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Florentz, C.; Sissler, M. Mitochondrial tRNA aminoacylation and human diseases. In Translation Mechanisms; Lapointe, J., Brakier-Gingras, L., Eds.; Landes Bioscience: Georgetown, TX, 2003; pp. 129–143. [Google Scholar]
- Bonnefond, L.; Frugier, M.; Giegé, R.; Rudinger-Thirion, J. Human mitochondrial TyrRS disobeys the tyrosine idenity rules. RNA 2005, 11, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, L.; Giegé, R.; Rudinger-Thirion, J. Evolution of the tRNATyr/TyrRS aminoacylation systems. Biochimie 2005, 87, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, L.; Frugier, M.; Touzé, E.; Lorber, B.; Florentz, C.; Giegé, R.; Sauter, C.; Rudinger-Thirion, J. Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features. Structure 2007, 15, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.L.; Tao, Z.J.; Jia, J.; He, X.X.; Jin, Y.X. Species-specific aminoacylation of Oryza sativa mitochondrial tRNATrp. Chinese Sci. Bull. 2006, 51, 824–829. [Google Scholar] [CrossRef]
- Shimada, N.; Suzuki, T.; Watanabe, K. Dual mode of recognition of two isoacceptor tRNAs by mammalian mitochondrial seryl-tRNA synthetase. J. Biol. Chem. 2001, 276, 46770–46778. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.R.; Saks, M.E. Contributions of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase: A kinetic analysis using model rna substrates. Nucleic Acids Res. 1993, 21, 4467–4475. [Google Scholar]
- Waeschenbach, A.; Porter, J.S.; Hughes, R.N. Molecular variability in the Celleporella hyalina (Bryozoa; Cheilostomata) species complex: Evidence for cryptic speciation from complete mitochondrial genomes. Mol. Biol. Rep. 2012, 39, 8601–8614. [Google Scholar] [CrossRef] [PubMed]
- Lovato, M.A.; Chihade, J.W.; Schimmel, P. Translocation within the acceptor helix of a major tRNA identity determinant. EMBO J. 2001, 20, 4846–4853. [Google Scholar] [CrossRef] [PubMed]
- Chihade, J.W.; Hayashibara, K.; Shiba, K.; Schimmel, P. Strong selective pressure to use G:U to mark an RNA acceptor stem for alanine. Biochemistry 1998, 37, 9193–9202. [Google Scholar] [CrossRef] [PubMed]
- Aphasizhev, R.; Senger, B.; Rengers, J.U.; Sprinzl, M.; Walter, P.; Nussbaum, G.; Fasiolo, F. Conservation in evolution for a small monomeric phenylalanyl-tRNA synthetase of the tRNAPhe recognition nucleotides and initial aminoacylation site. Biochemistry 1996, 35, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Tinkle-Peterson, E.; Uhlenbeck, O.C. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry 1992, 31, 10380–10389. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Lapointe, J. Transfer RNA aminoacylation and modified nucleosides. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution; Grosjean, H., Ed.; Landes Bioscience: Georgetown, TX, USA, 2009; pp. 475–492. [Google Scholar]
- Renaud, M.; Ehrlich, R.; Bonnet, J.; Remy, P. Lack of correlation between affinity of the tRNA for the aminoacyl-tRNA synthetase and aminoacylation capacity as studied with modified tRNAPhe. Eur. J. Biochem. 1979, 100, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.M.; Motorin, Y.A.; Wolfson, A.D.; Gladilin, K.L. Anticodon-dependent aminoacylation of RNA minisubstrate by lysyl-tRNA synthetase. FEBS Lett. 1992, 314, 256–258. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salinas-Giegé, T.; Giegé, R.; Giegé, P. tRNA Biology in Mitochondria. Int. J. Mol. Sci. 2015, 16, 4518-4559. https://doi.org/10.3390/ijms16034518
Salinas-Giegé T, Giegé R, Giegé P. tRNA Biology in Mitochondria. International Journal of Molecular Sciences. 2015; 16(3):4518-4559. https://doi.org/10.3390/ijms16034518
Chicago/Turabian StyleSalinas-Giegé, Thalia, Richard Giegé, and Philippe Giegé. 2015. "tRNA Biology in Mitochondria" International Journal of Molecular Sciences 16, no. 3: 4518-4559. https://doi.org/10.3390/ijms16034518
APA StyleSalinas-Giegé, T., Giegé, R., & Giegé, P. (2015). tRNA Biology in Mitochondria. International Journal of Molecular Sciences, 16(3), 4518-4559. https://doi.org/10.3390/ijms16034518





