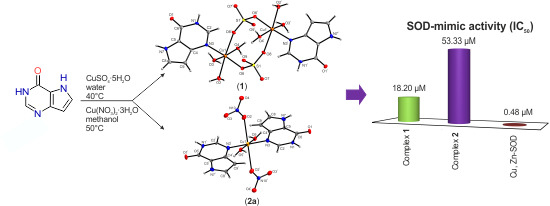
Structural Diversity of Copper(II) Complexes with 9-Deazahypoxanthine and Their in Vitro SOD-Like Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and General Properties

2.2. TG and DTA Thermal Analyses
2.3. Electrospray Ionization (ESI) Mass Spectrometry
2.4. Infrared Spectroscopy
2.5. UV-Vis Spectroscopy
2.6. Single Crystal X-ray Analysis
| Compound | 1 | 2a |
|---|---|---|
| Empirical formula | C12H22Cu2N6O16S2 | C12H14Cu1N8O10 |
| Formula weight | 697.56 | 493.85 |
| Temperature (K) | 100(2) | 100(2) |
| Wavelength (Å) | 0.71073 | 0.71073 |
| Crystal system | Triclinic | Triclinic |
| Space group | P-1 | P-1 |
| a (Å) | 6.8164 (2) | 8.6756 (5) |
| b (Å) | 8.9268 (3) | 8.9906 (5) |
| c (Å) | 9.2297 (3) | 13.0055 (7) |
| α (°) | 80.073 (3) | 89.127 (5) |
| β (°) | 81.919 (3) | 74.514 (5) |
| γ (°) | 85.943 (3) | 61.386 (5) |
| V (Å3) | 547.08 (3) | 850.55 (8) |
| Z, Dc (g·cm−3) | 1, 2.117 | 2, 1.924 |
| F (000) | 354 | 500 |
| θ range for data collection (°) | 3.02 ≤ θ ≤ 25.00 | 2.89 ≤ θ ≤ 25.00 |
| Reflections collected/unique | 5129/1920 (R(int) = 0.0211) | 7572/2991 (R(int) = 0.0302) |
| Data/restraints/parameters | 1920/4/190 | 2991/0/242 |
| Goodness-of-fit on F2 | 1.078 | 1.246 |
| Final R indices (I > 2σ(I)) | R1 = 0.0306, wR2 = 0.0823 | R1 = 0.0842, wR2 = 0.2433 |
| R indices (all data) | R1 = 0.0393, wR2 = 0.0862 | R1 = 0.0879, wR2 = 0.2444 |
| Largest peak and hole (e·Å−3) | 0.789, −0.582 | 1.758, −0.787 |
| 1 | |||
| Cu(1)–O(4) | 1.977(2) | O(4)–Cu(1)–O(3) | 169.32(9) |
| Cu(1)–O(6) | 1.980(2) | O(4)–Cu(1)–O(2) | 91.56(9) |
| Cu(1)–O(3) | 1.981(2) | O(6)–Cu(1)–N(3) | 175.19(9) |
| Cu(1)–O(2) | 2.329(2) | N(3)–Cu(1)–O(2) | 91.75(9) |
| Cu(1)–N(3) | 2.018(3) | O(6)–Cu(1)–O(8) i | 94.02(8) |
| Cu(1)–O(8) i | 2.428(2) | O(2)–Cu(1)–O(8) i | 174.83(7) |
| 2a/2aA | |||
| Cu(1)–O(5)/Cu(1)–O(5) i | 1.975(6) | N(3)–Cu(1)–N(3) i/N(3A)–Cu(2)–N(3A) ii | 179.999(1) |
| Cu(1)–N(3)/Cu(1)–N(3) i | 2.007(7) | O(2)–Cu(1)–O(2) i/O(2A)–Cu(2)–O(2A) ii | 180.000(1) |
| Cu(1)–O(2)/Cu(1)–O(2) i | 2.524(6) | O(5) i–Cu(1)–O(5)/O(5A)–Cu(2)–O(5A) ii | 180.000(1) |
| Cu(2)–O(5A)/Cu(2)–O(5A) ii | 1.994(6) | O(5)–Cu(1)–N(3)/O(5A)–Cu(2)–N(3A) | 90.2(3)/89.0(3) |
| Cu(2)–N(3A)/Cu(2)–N(3A) ii | 2.031(8) | O(5)–Cu(1)–N(3) i/O(5A) ii–Cu(2)–N(3A) | 89.8(3)/91.0(3) |
| Cu(2)–O(2A)/Cu(2)–O(2A) ii | 2.464(8) | O(5)–Cu(1)–O(2)/O(5A)–Cu(2)–O(2A) | 81.0(2)/81.5(2) |
2.6.1. X-ray Structure of [{Cu(9dhx)(H2O)3}2(µ-SO4)2] (1)



2.6.2. X-ray Structure of [Cu(9dhx)2(H2O)2(NO3)2] (2a)



2.7. In Vitro SOD-Like Activity
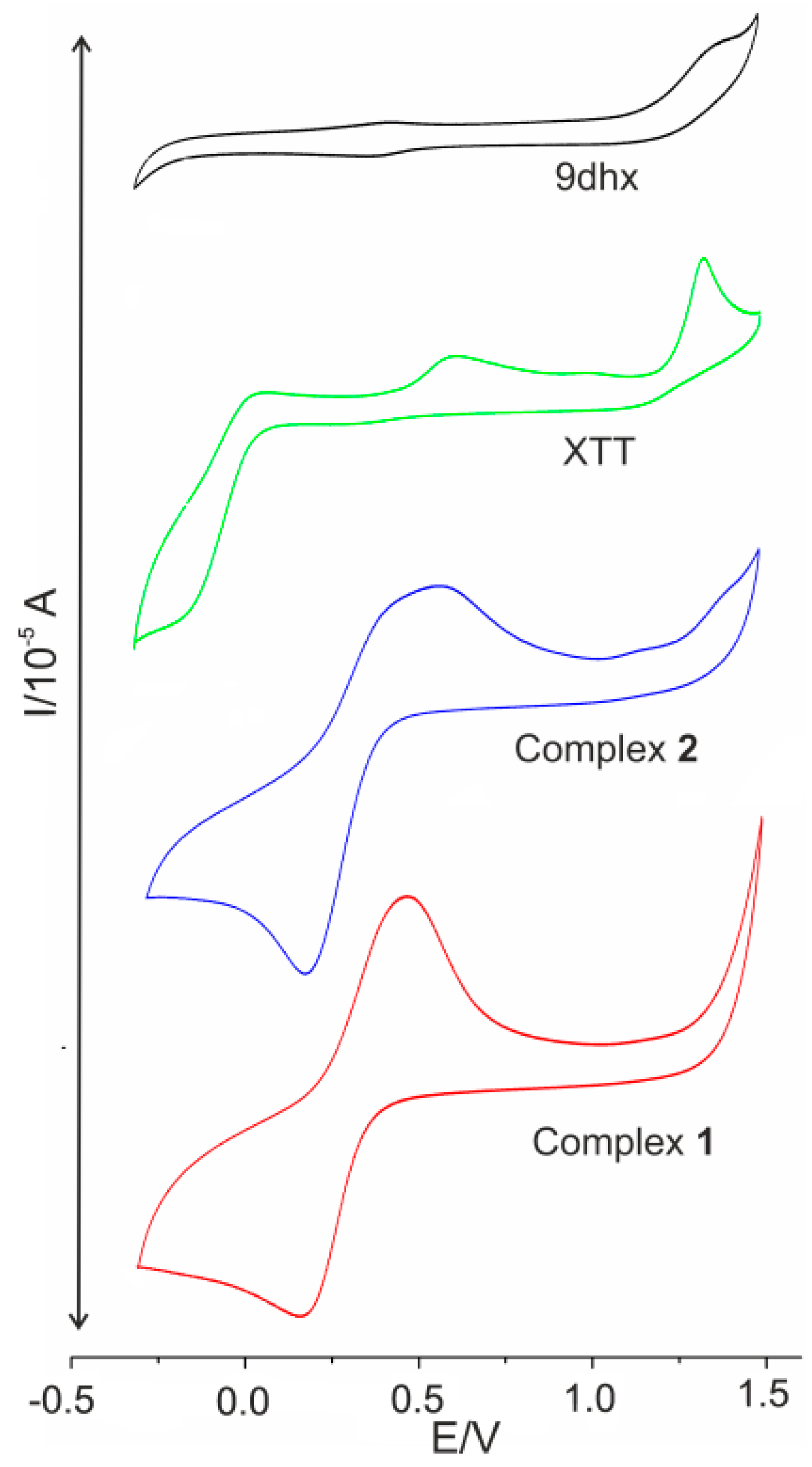
2.8. Cyclic Voltammetry
| Compound | DMSO | ||
|---|---|---|---|
| Eox * | Ered * | Ipa, Ipc | |
| 1 | 306 | 303 | −29, 23 |
| 2 | 325 | 326 | −22, 29 |
| XTT | Eox1 = 1326 Eox2 = 614 | Ered1 = −137 | |
3. Experimental Section
3.1. Starting Materials
3.2. Synthesis of [{Cu(9dhx)(H2O)3}2(µ-SO4)2] (1)
3.3. Synthesis of [Cu(9dhx)2(H2O)2(NO3)2]·H2O (2)
3.4. Methods of Characterization
3.5. SOD-Like Activity Testing
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Solomon, E.I.; Heppner, D.E.; Johnston, E.M.; Ginsbach, J.W.; Cirera, J.; Qayyum, M.; Kieber-Emmons, M.T.; Kjaergaard, C.H.; Hadt, R.G.; Tian, L. Copper active sites in biology. Chem. Rev. 2014, 114, 3659–3853. [Google Scholar] [CrossRef] [PubMed]
- Chrichton, R.R. Biological Inorganic Chemistry: A New Introduction to Molecular Structure and Function, 2nd ed.; Elsevier: Oxford, UK, 2012. [Google Scholar]
- Linder, M.C. Biochemistry of Copper; Plenum Press: New York, NY, USA, 1991. [Google Scholar]
- Fridovich, I. Superoxide dismutases. Annu. Rev. Biochem. 1975, 44, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Veni, K.G.; Usha, B.; Kumar, D.M.; Rao, T.R. Alterations in serum SOD and CAT levels in patients with breast cancer. J. Exp. Sci. 2011, 2, 58–60. [Google Scholar]
- Salminen, A.; Ojala, J.; Kaarniranta, K.; Kauppinen, A. Mitochondrial dysfunction and oxidative stress activate inflammasomes: Impact on the aging process and age-related diseases. Cell. Mol. Life Sci. 2012, 69, 2999–3013. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, J.J.A.; Teunissen, C.E.; de Vries, H.E.; Dijkstra, C.D. Macrophages and neurodegeneration. Brain Res. Rev. 2005, 48, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Margaill, I.; Plotkine, M.; Lerouet, D. Antioxidant strategies in the treatment of stroke. Free Radic. Biol. Med. 2005, 39, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Jay, D.; Hitomi, H.; Griendling, K.K. Oxidative stress and diabetic cardiovascular complications. Free Radic. Biol. Med. 2006, 40, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.; Shiloh, M.U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 2000, 97, 8841–8848. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.C.; Kammouni, W.; Zherebitskaya, E.; Fernyhough, P. Role of oxidative stress in rabies virus infection of adult mouse dorsal root ganglion neurons. J. Virol. 2010, 84, 4697–4705. [Google Scholar] [CrossRef] [PubMed]
- Gupta-Elera, G.; Garrett, A.R.; Robison, R.A.; O’Neill, K.L. The role of oxidative stress in prostate cancer. Eur. J. Cancer Prev. 2012, 21, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Belda, R.; Blasco, S.; Verdejo, B.; Jiménez, H.R.; Doménech-Carbó, A.; Soriano, C.; Latorre, J.; Terencio, C.; García-España, E. Homo- and heterobinuclear Cu2+ and Zn2+ complexes of abiotic cyclic hexaazapyridinocyclophanes as SOD mimics. Dalton Trans. 2013, 42, 11194–11204. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, Z.A.; Shahid, M.; Khalid, M.; Kumar, S. Antimicrobial and SOD activities of novel transition metal ternary complexes of iminodiacetic acid containing α-diimine as auxiliary ligand. Eur. J. Med. Chem. 2009, 44, 2517–2522. [Google Scholar] [CrossRef] [PubMed]
- Vančo, J.; Švajlenová, O.; Račanská, E.; Muselík, J.; Valentová, J. Antiradical activity of different copper(II) Schiff base complexes and their effect on alloxan-induced diabetes. J. Trace Elem. Med. Biol. 2004, 18, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Štarha, P.; Trávníček, Z.; Herchel, R.; Popa, I.; Suchý, P.; Vančo, J. Dinuclear copper(II) complexes containing 6-(benzylamino)purines as bridging ligands: Synthesis, characterization, and in vitro and in vivo antioxidant activities. J. Inorg. Biochem. 2009, 103, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Klanicová, A.; Trávníček, Z.; Vančo, J.; Popa, I.; Šindelář, Z. Dinuclear copper(II) perchlorate complexes with 6-(benzylamino)purine derivatives: Synthesis, X-ray structure, magnetism and antiradical activity. Polyhedron 2010, 29, 2582–2589. [Google Scholar] [CrossRef]
- Novotná, R.; Trávníček, Z.; Herchel, R. SOD-Mimic Cu(II) dimeric complexes involving kinetin and its derivative: Preparation and characterization. Bioinorg. Chem. Appl. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Bantia, S.; Miller, P.J.; Parker, C.D.; Ananth, S.L.; Horn, L.L.; Kilpatrick, J.M.; Morris, P.E.; Hutchison, T.L.; Montgomery, J.A.; Sandhu, J.S. Purine phosphorylase inhibitor BCX-1777 (Immucillin-H)–a novel potent and orally active immunosuppressive agent. Int. Immunopharmacol. 2001, 1, 1199–1210. [Google Scholar] [CrossRef]
- Dummer, R.; Duvic, M.; Scarisbrick, J.; Olsen, E.A.; Rozati, S.; Eggmann, N.; Oldinger, S.M.; Hutchinson, K.; Geskin, L.; Illidge, T.M.; et al. Final results of a multicenter phase II study of the purine nucleoside phosphorylase (PNP) inhibitor forodesine in patients with advanced cutaneous t-cell lymphomas (CTCL) (Mycosis fungoides and Sézary syndrome). Ann. Oncol. 2014, 25, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Gáliková, J.; Trávníček, Z. Effect of different reaction conditions on the structural diversity of zinc (II) complexes with 9-deazahypoxanthine. Polyhedron 2014, 79, 269–276. [Google Scholar] [CrossRef]
- Vančo, J.; Gáliková, J.; Hošek, J.; Dvořák, Z.; Paráková, L.; Trávníček, Z. Gold(I) complexes of 9-deazahypoxanthine as selective antitumor and anti-inflammatory agents. PLoS ONE 2014, 9, e109901. [Google Scholar] [CrossRef] [PubMed]
- Gáliková, J.; Hošek, J.; Trávníček, Z. Synthesis, X-ray crystal structure and biological evaluation of zinc(II)-dichlorido complexes with 9-deazahypoxathine derivatives. Inorg. Chim. Acta 2015, 434, 67–73. [Google Scholar] [CrossRef]
- Novotná, R.; Trávníček, Z. Infinite ladder-like chains organized into a three-dimensional zigzag supra-molecular architecture in 9-deazahypoxanthine. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 2013, C69, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Kamath, V.P.; Juarez-Brambila, J.J.; Morris, C.B.; Winslow, C.D.; Morris, P.E., Jr. Development of a practical synthesis of a purine nucleoside phosphorylase inhibitor: BCX-4208. Org. Process Res. Dev. 2009, 13, 928–932. [Google Scholar] [CrossRef]
- Geary, W.J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Nakamoto, K.; Margoshes, M.; Rundle, R.E. Stretching frequencies as a function of distances in hydrogen bonds. J. Am. Chem. Soc. 1955, 77, 6480–6486. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds, Part B: Applications in Coordination, Orgametallic and Bioinorganic Chemistry, 5th ed.; Wiley: New York, NY, USA, 1997. [Google Scholar]
- Solomon, E.I.; Lever, A.B.P. Inorganic Electronic Structure and Spectroscopy, Applications and Case Studies; Wiley: New York, NY, USA, 1999; Volume 2. [Google Scholar]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. Sect. B Struct. Sci. 2002, 58, 380–388. [Google Scholar] [CrossRef]
- Jitsukawa, K.; Harata, M.; Arii, H.; Sakurai, H.; Masuda, H. SOD activities of the copper complexes with tripodal polypyridylamine ligands having a hydrogen bonding site. Inorg. Chim. Acta 2001, 324, 108–116. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Sun, D.L.; Tao, J.; Chen, L.Q.; Huang, Y.F.; Li, Y.K.; Cheng, Y. Synthesis, crystal structure, and SOD-like activity of two copper(II) complexes with hydroxymethyl pendants. J. Coord. Chem. 2014, 67, 2393–2404. [Google Scholar] [CrossRef]
- Klanicová, A.; Houck, J.D.; Baran, P.; Trávníček, Z. Synthesis, X-ray structures, properties and SOD-like activity of ternary copper(II) complexes showing the N4O2 coordination with a combination of monodentate and bidentate N-donor ligands. Inorg. Chim. Acta 2012, 384, 47–53. [Google Scholar] [CrossRef]
- Bóka, B.; Myari, A.; Sóvágó, I.; Hadjiliadis, N. Copper(II) and zinc(II) complexes of the peptides Ac-HisValHis-NH2 and Ac-HisValGlyAsp-NH2 related to the active site of the enzyme CuZnSOD. J. Inorg. Biochem. 2004, 98, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Zoski, C.G. Handbook of Electrochemistry, 1st ed.; Elsevier: Oxford, UK, 2007. [Google Scholar]
- Oxford Diffraction. CrysAlis RED and CrysAlis CCD Software (Ver. 1.171.33.52); Oxford Diffraction Ltd.: Abingdon, Oxfordshire, UK, 2009. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, K. DIAMOND; Release 3.2i; Crystal Impact GbR: Bonn, Germany, 2011. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gáliková, J.; Trávníček, Z. Structural Diversity of Copper(II) Complexes with 9-Deazahypoxanthine and Their in Vitro SOD-Like Activity. Int. J. Mol. Sci. 2015, 16, 15954-15970. https://doi.org/10.3390/ijms160715954
Gáliková J, Trávníček Z. Structural Diversity of Copper(II) Complexes with 9-Deazahypoxanthine and Their in Vitro SOD-Like Activity. International Journal of Molecular Sciences. 2015; 16(7):15954-15970. https://doi.org/10.3390/ijms160715954
Chicago/Turabian StyleGáliková, Jana, and Zdeněk Trávníček. 2015. "Structural Diversity of Copper(II) Complexes with 9-Deazahypoxanthine and Their in Vitro SOD-Like Activity" International Journal of Molecular Sciences 16, no. 7: 15954-15970. https://doi.org/10.3390/ijms160715954
APA StyleGáliková, J., & Trávníček, Z. (2015). Structural Diversity of Copper(II) Complexes with 9-Deazahypoxanthine and Their in Vitro SOD-Like Activity. International Journal of Molecular Sciences, 16(7), 15954-15970. https://doi.org/10.3390/ijms160715954