Pyrosequencing-Based Assays for Rapid Detection of HER2 and HER3 Mutations in Clinical Samples Uncover an E332E Mutation Affecting HER3 in Retroperitoneal Leiomyosarcoma
Abstract
:1. Introduction
2. Results
3. Discussion
| Tumor Type | Specimens | HER2 | HER3 | |||
|---|---|---|---|---|---|---|
| Category | Sample size (n) | Mutants (n) | aa Change | wt/mutant codon | Mutants (n) | aa Change |
| NSCLC | 10 | 0 | - | - | 0 | - |
| Breast Ca | 2 | 0 | - | - | 0 | - |
| CRC | 34 | 1 | L755M | TTG/ATG | 1 | D297Y |
| 1 | V842I | GTA/ATA | ||||
| Bladder Urothelial Cell Ca | 11 | 0 | - | - | 1 | E332K |
| Epithelial Ovarian Ca | 7 | 1 | V842I | GTA/ATA | 0 | - |
| Endometrial Ca | 3 | 0 | - | - | 0 | - |
| Cervical Squamous Cell Ca | 1 | 0 | - | - | 0 | - |
| Gastric Ca | 9 | 0 | - | - | 1 | D297Y |
| Pancreatic Ca | 2 | 1 | V842I | GTA/ATA | 0 | - |
| Soft tissue sarcoma | 3 | 0 | - | - | 1 | E332E |
| RCC | 2 | 0 | - | - | 0 | - |
| Melanoma | 1 | 0 | - | - | 0 | - |
| Cutaneous SCC | 1 | 0 | - | - | 0 | - |
| Total | 86 | 4 | 4 | |||
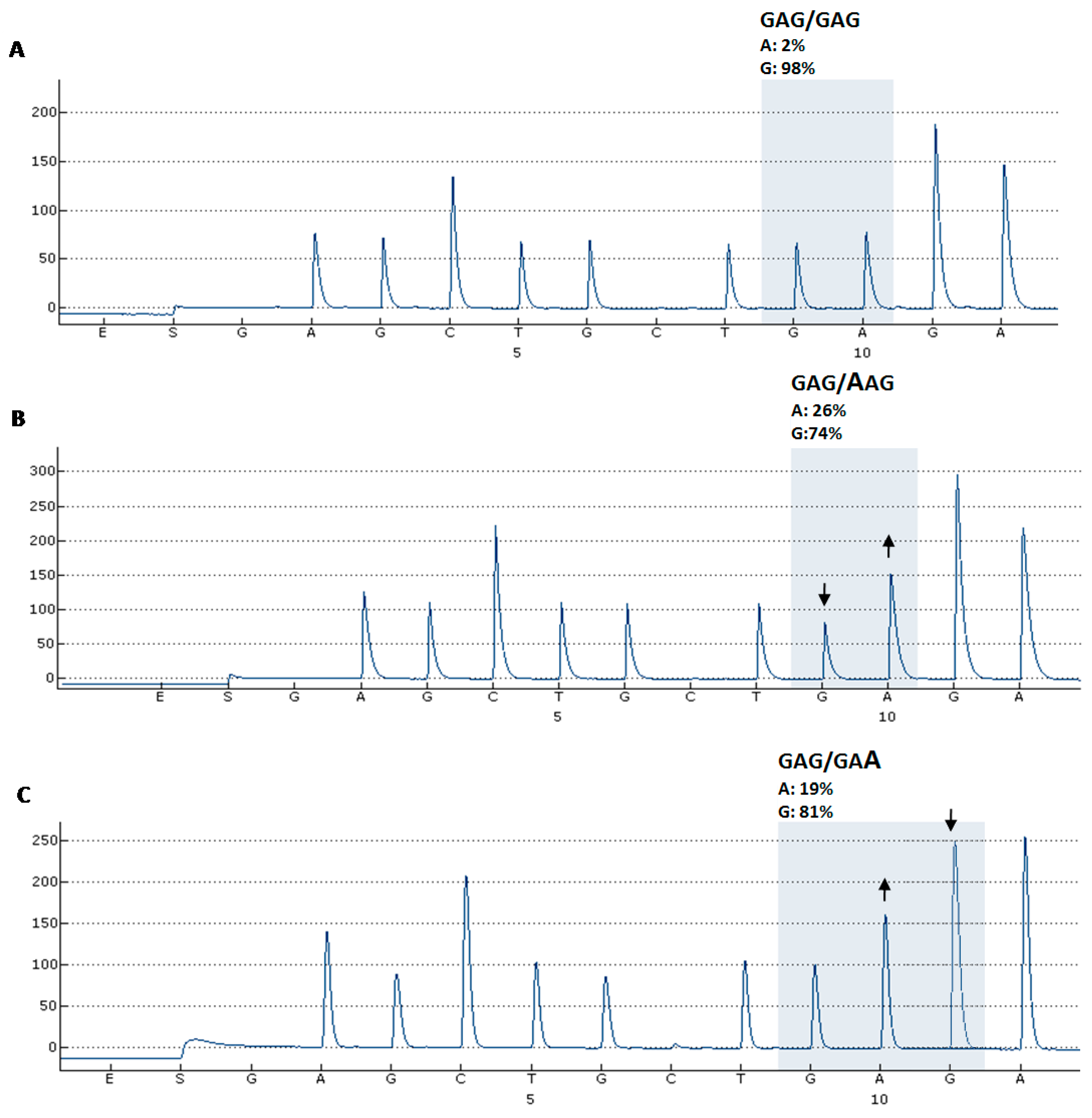
4. Experimental Section
4.1. Clinical Samples
4.2. Nucleic Acid Extraction and cDNA Synthesis
4.3. PCR Primers Design and PCR Amplification
4.4. Electrophoresis
4.5. Pyrosequencing Primers Design and Pyrosequencing Assay

| Target | Assay | NDO |
|---|---|---|
| HER2 | S310 | GTGATCTGTCACT |
| R678 | TAGCAGTCAGAG | |
| L755 | GAGTCGATCGAG | |
| V777 | CGATGCATGCT | |
| V842 | AGCTCAGTACGACAG | |
| HER3 | M91 | TCACTCGATCGA |
| V104 | GCGCTGACTACGTAGC | |
| D297 | CGTATCGACA | |
| E332 | GAGCTGCTGAGA |
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hynes, N.E.; Lane, H.A. ErbB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Burgess, A.W.; Cho, H.S.; Eigenbrot, C.; Ferguson, K.M.; Garrett, T.P.; Leahy, D.J.; Lemmon, M.A.; Sliwkowski, M.X.; Ward, C.W.; Yokoyama, S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell 2003, 12, 541–552. [Google Scholar] [CrossRef]
- Baselga, J.; Swain, S.M. Novel anticancer targets: Revisiting ErbB2 and discovering ErbB3. Nat. Rev. Cancer 2009, 9, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.; Kavuri, S.M.; Searleman, A.C.; Shen, W.; Shen, D.; Koboldt, D.C.; Monsey, J.; Goel, N.; Aronson, A.B.; Li, S.; et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013, 3, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Xiao, Y.; Joshi, V.; Yeap, B.Y.; Cioffredi, L.A.; Jackman, D.M.; Lee, C.; Janne, P.A.; Lindeman, N.I. EGFR mutation is a better predictor of response to tyrosine kinase inhibitors in non-small cell lung carcinoma than FISH, CISH, and immunohistochemistry. Am. J. Clin. Pathol. 2010, 133, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, T.H.; Chakraborty, S.; Paul, P. A cross talk between codon usage bias in human oncogenes. Bioinformation 2014, 10, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Wheler, J.J.; Parker, B.A.; Lee, J.J.; Atkins, J.T.; Janku, F.; Tsimberidou, A.M.; Zinner, R.; Subbiah, V.; Fu, S.; Schwab, R.; et al. Unique molecular signatures as a hallmark of patients with metastatic breast cancer: Implications for current treatment paradigms. Oncotarget 2014, 5, 2349–2354. [Google Scholar] [PubMed]
- Herter-Sprie, G.S.; Greulich, H.; Wong, K.K. Activating mutations in ErbB2 and their impact on diagnostics and treatment. Front. Oncol. 2013, 3, 86. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, B.S.; Kljavin, N.M.; Stawiski, E.W.; Chan, E.; Parikh, C.; Durinck, S.; Chaudhuri, S.; Pujara, K.; Guillory, J.; Edgar, K.A.; et al. Oncogenic ErbB3 mutations in human cancers. Cancer Cell 2013, 23, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Sundstrom, M.; Edlund, K.; Lindell, M.; Glimelius, B.; Birgisson, H.; Micke, P.; Botling, J. KRAS analysis in colorectal carcinoma: Analytical aspects of pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer 2010, 10, 660. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Bahleda, R.; Tolaney, S.M.; Kwak, E.L.; Cleary, J.M.; Pandya, S.S.; Hollebecque, A.; Abbas, R.; Ananthakrishnan, R.; Berkenblit, A.; et al. Phase I study of neratinib in combination with temsirolimus in patients with human epidermal growth factor receptor 2-dependent and other solid tumors. J. Clin. Oncol. 2014, 32, 68–75. [Google Scholar] [CrossRef] [PubMed]
- cBioPortal for cancer genomics. Available online: http://www.cbioportal.org/ (accessed on 12 August 2015).
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Holbrook, J.D.; Parker, J.S.; Gallagher, K.T.; Halsey, W.S.; Hughes, A.M.; Weigman, V.J.; Lebowitz, P.F.; Kumar, R. Deep sequencing of gastric carcinoma reveals somatic mutations relevant to personalized medicine. J. Transl. Med. 2011, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Cosmic: Catalogue of somatic mutations in cancer. Available online: http://cancer.sanger.ac.uk/cancergenome/projects/cosmic (accessed on 12 August 2015).
- NCBI (Nacional center for biotechnology information). dbSNP: Database of single nucleotide polymorphisms (SNPs). Available online: http://www.ncbi.nlm.nih.gov/snp (accessed on 12 August 2015).
- UniProt: Universal protein resource. Available online: http://www.uniprot.org/uniprot/P21860 (accessed on 12 August 2015).
- Zheng, S.; Kim, H.; Verhaak, R.G. Silent mutations make some noise. Cell 2014, 156, 1129–1131. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Minana, B.; Valcarcel, J.; Gabaldon, T.; Lehner, B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 2014, 156, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Prober, J.M.; Trainor, G.L.; Dam, R.J.; Hobbs, F.W.; Robertson, C.W.; Zagursky, R.J.; Cocuzza, A.J.; Jensen, M.A.; Baumeister, K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science 1987, 238, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Righi, L.; Cuccurullo, A.; Vatrano, S.; Cappia, S.; Giachino, D.; de Giuli, P.; Ardine, M.; Novello, S.; Volante, M.; Scagliotti, G.V.; et al. Detection and characterization of classical and “uncommon” exon 19 epidermal growth factor receptor mutations in lung cancer by pyrosequencing. BMC Cancer 2013, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Nacional center for biotechnology information. Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 12 August 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Alonso, P.; Chamizo, C.; Moreno, V.; Madoz-Gúrpide, J.; Carvajal, N.; Daoud, L.; Zazo, S.; Martín-Aparicio, E.; Cristóbal, I.; Rincón, R.; et al. Pyrosequencing-Based Assays for Rapid Detection of HER2 and HER3 Mutations in Clinical Samples Uncover an E332E Mutation Affecting HER3 in Retroperitoneal Leiomyosarcoma. Int. J. Mol. Sci. 2015, 16, 19447-19457. https://doi.org/10.3390/ijms160819447
González-Alonso P, Chamizo C, Moreno V, Madoz-Gúrpide J, Carvajal N, Daoud L, Zazo S, Martín-Aparicio E, Cristóbal I, Rincón R, et al. Pyrosequencing-Based Assays for Rapid Detection of HER2 and HER3 Mutations in Clinical Samples Uncover an E332E Mutation Affecting HER3 in Retroperitoneal Leiomyosarcoma. International Journal of Molecular Sciences. 2015; 16(8):19447-19457. https://doi.org/10.3390/ijms160819447
Chicago/Turabian StyleGonzález-Alonso, Paula, Cristina Chamizo, Víctor Moreno, Juan Madoz-Gúrpide, Nerea Carvajal, Lina Daoud, Sandra Zazo, Ester Martín-Aparicio, Ion Cristóbal, Raúl Rincón, and et al. 2015. "Pyrosequencing-Based Assays for Rapid Detection of HER2 and HER3 Mutations in Clinical Samples Uncover an E332E Mutation Affecting HER3 in Retroperitoneal Leiomyosarcoma" International Journal of Molecular Sciences 16, no. 8: 19447-19457. https://doi.org/10.3390/ijms160819447
APA StyleGonzález-Alonso, P., Chamizo, C., Moreno, V., Madoz-Gúrpide, J., Carvajal, N., Daoud, L., Zazo, S., Martín-Aparicio, E., Cristóbal, I., Rincón, R., García-Foncillas, J., & Rojo, F. (2015). Pyrosequencing-Based Assays for Rapid Detection of HER2 and HER3 Mutations in Clinical Samples Uncover an E332E Mutation Affecting HER3 in Retroperitoneal Leiomyosarcoma. International Journal of Molecular Sciences, 16(8), 19447-19457. https://doi.org/10.3390/ijms160819447






