Isolation of Endogenously Assembled RNA-Protein Complexes Using Affinity Purification Based on Streptavidin Aptamer S1
Abstract
:1. Introduction
2. Results
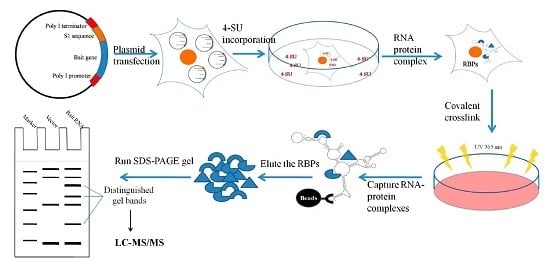
2.1. Design and Expression of Plasmid-Based S1-Aptamer-Tagged 5′–3′ UTR RNA

2.2. Isolating Endogenously-Assembled DENV 5′–3′U-S1 RNPs

| Description | Protein Score | Protein Mass (kDa) | Coverage (%) | Unique Peptide |
|---|---|---|---|---|
| PHF6 | 342.27 | 42.4 | 7.41 | 3 |
| RBMXL1 | 463.26 | 42.2 | 19.98 | 6 |
| HNRNPD | 478.9 | 38.6 | 21.84 | 6 |
| SNRPB | 136.3 | 24.8 | 5.842 | 2 |
| RPS8 | 427.5 | 24.5 | 27.88 | 5 |
2.3. Involvement of the Newly Identified 5′–3′ UTR RBP RPS8 in DENV RNA Replication in Cells


3. Discussion
4. Experimental Section
4.1. Plasmid Construction
4.2. Cell Culture, Plasmid Transfection, Crosslinking, and Cell Lysis
4.3. DENV-2 Amplification and Infection
4.4. Reverse Transcript PCR (RT-PCR) and Relative Quantitative Real-Time PCR (RT-qPCR) Analysis
4.5. RNA Pull-down Assay
4.6. SDS-PAGE and Western Blotting
4.7. Mass Spectrometry
4.8. Immunofluorescence Assay and Confocal Microscopy
4.9. RNA Immunoprecipitation
4.10. Plaque Assay
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, Z.; Nagy, P.D. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011, 8, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Tuplin, A. Diverse roles and interactions of RNA structures during the replication of positive-stranded RNA viruses of humans and animals. J. Gen. Virol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999, 37, 151–174. [Google Scholar] [CrossRef] [PubMed]
- Windbichler, N.; Schroeder, R. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nat. Protoc. 2006, 1, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Bachler, M.; Schroeder, R.; von Ahsen, U. StreptoTag: A novel method for the isolation of RNA-binding proteins. RNA 1999, 5, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Hartmuth, K.; Vornlocher, H.P.; Luhrmann, R. Tobramycin affinity tag purification of spliceosomes. Methods Mol. Biol. 2004, 257, 47–64. [Google Scholar] [PubMed]
- Srisawat, C.; Engelke, D.R. Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA 2001, 7, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Czaplinski, K.; Kocher, T.; Schelder, M.; Segref, A.; Wilm, M.; Mattaj, I.W. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-β-related mRNA during oogenesis. Dev. Cell 2005, 8, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.; Grskovic, M.; Strein, C.; Beckmann, K.; Niggeweg, R.; Abaza, I.; Gebauer, F.; Wilm, M.; Hentze, M.W. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: Translational repression for dosage compensation. Genes Dev. 2006, 20, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.M.; Gunaratne, J.; Garcia-Blanco, M.A. Identification of dengue RNA binding proteins using RNA chromatography and quantitative mass spectrometry. Methods Mol. Biol. 2014, 1138, 253–270. [Google Scholar] [PubMed]
- Nonne, N.; Ameyar-Zazoua, M.; Souidi, M.; Harel-Bellan, A. Tandem affinity purification of miRNA target mRNAs (TAP-Tar). Nucleic Acids Res. 2010, 38. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Butter, F.; Scheibe, M.; Morl, M.; Mann, M. Unbiased RNA-protein interaction screen by quantitative proteomics. Proc. Natl. Acad. Sci. USA 2009, 106, 10626–10631. [Google Scholar] [CrossRef] [PubMed]
- Hogg, J.R.; Collins, K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 2007, 13, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Hogg, J.R.; Collins, K. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal RNA quality control. Genes Dev. 2007, 21, 3067–3072. [Google Scholar] [CrossRef] [PubMed]
- Hogg, J.R.; Goff, S.P. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 2010, 143, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Lemay, V.; Hossain, A.; Osheim, Y.N.; Beyer, A.L.; Dragon, F. Identification of novel proteins associated with yeast snR30 small nucleolar RNA. Nucleic Acids Res. 2011, 39, 9659–9670. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Haurwitz, R.E.; Apffel, A.; Zhou, K.; Smart, B.; Wenger, C.D.; Laderman, S.; Bruhn, L.; Doudna, J.A. RNA-protein analysis using a conditional CRISPR nuclease. Proc. Natl. Acad. Sci. USA 2013, 110, 5416–5421. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.R. Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Res. 1979, 6, 715–732. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef] [PubMed]
- RNAfold Web Server. Available online: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi (accessed on 11 December 2013).
- UniProt. Available online: http://www.uniprot.org/ (accessed on 18 July 2014).
- Wang, J.; Leung, J.W.; Gong, Z.; Feng, L.; Shi, X.; Chen, J. PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J. Biol. Chem. 2013, 288, 3174–3183. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Mosbrook, D.M.; Tagle, D.A. Genomic organization and expression analysis of mouse kynurenine aminotransferase II, a possible factor in the pathophysiology of Huntington’s disease. Mamm. Genome 1999, 10, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Grosset, C.; Chen, C.Y.; Xu, N.; Sonenberg, N.; Jacquemin-Sablon, H.; Shyu, A.B. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 2000, 103, 29–40. [Google Scholar] [CrossRef]
- Chari, A.; Golas, M.M.; Klingenhager, M.; Neuenkirchen, N.; Sander, B.; Englbrecht, C.; Sickmann, A.; Stark, H.; Fischer, U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 2008, 135, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Jonson, L.; Vikesaa, J.; Krogh, A.; Nielsen, L.K.; Hansen, T.; Borup, R.; Johnsen, A.H.; Christiansen, J.; Nielsen, F.C. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteom. 2007, 6, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Watanabe, T.; Ito, H.; Watanabe, S.; Goto, H.; Gao, P.; Hughes, M.; Perez, D.R.; Donis, R.; Hoffmann, E.; et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA. 1999, 96, 9345–9350. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Gamarnik, A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009, 139, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.C.; Munschauer, M.; et al. PAR-CliP—A method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp. 2010, 41, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Reed, R. Purification of functional RNA-protein complexes using MS2-MBP. Curr. Protoc. Mol. Biol. 2003, 27, 1–27. [Google Scholar]
- Bardwell, V.J.; Wickens, M. Purification of RNA and RNA-protein complexes by an R17 coat protein affinity method. Nucleic Acids Res. 1990, 18, 6587–6594. [Google Scholar] [CrossRef] [PubMed]
- Tsai, B.P.; Wang, X.; Huang, L.; Waterman, M.L. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Keryer-Bibens, C.; Barreau, C.; Osborne, H.B. Tethering of proteins to RNAs by bacteriophage proteins. Biol. Cell 2008, 100, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Hook, B.; Bernstein, D.; Zhang, B.; Wickens, M. RNA-protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA 2005, 11, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J. RNA-protein interaction protocols. Preface. Methods Mol. Biol. 2008, 488, v–vii. [Google Scholar] [PubMed]
- Zielinski, J.; Kilk, K.; Peritz, T.; Kannanayakal, T.; Miyashiro, K.Y.; Eiriksdottir, E.; Jochems, J.; Langel, U.; Eberwine, J. In vivo identification of ribonucleoprotein-RNA interactions. Proc. Natl. Acad. Sci. USA. 2006, 103, 1557–1562. [Google Scholar] [CrossRef] [PubMed]
- Bung, C.; Bochkaeva, Z.; Terenin, I.; Zinovkin, R.; Shatsky, I.N.; Niepmann, M. Influence of the hepatitis C virus 3′-untranslated region on IRES-dependent and cap-dependent translation initiation. FEBS Lett. 2010, 584, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yuan, B.; Lu, N.; Facchinetti, V.; Liu, Y.J. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J. Immunol. 2011, 187, 4501–4508. [Google Scholar] [CrossRef] [PubMed]
- Gomila, R.C.; Martin, G.W.; Gehrke, L. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS ONE 2011, 6, e16687. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Hanley, K.A. RNA interference modulates replication of dengue virus in Drosophila melanogaster cells. BMC Microbiol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Paranjape, S.M.; Harris, E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007, 282, 30497–30508. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.; Ketner, G.; Levis, R.; Falgout, B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J. Virol. 1997, 71, 5366–5374. [Google Scholar] [PubMed]
- Pang, X.; Zhang, M.; Dayton, A.I. Development of Dengue virus type 2 replicons capable of prolonged expression in host cells. BMC Microbiol. 2001, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Lenarcic, E.M.; Landry, D.M.; Greco, T.M.; Cristea, I.M.; Thompson, S.R. Thiouracil cross-linking mass spectrometry: A cell-based method to identify host factors involved in viral amplification. J. Virol. 2013, 87, 8697–8712. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Y.; Yang, J.; Ye, W.; Wang, Y.; Ye, C.; Weng, D.; Gao, H.; Zhang, F.; Xu, Z.; Lei, Y. Isolation of Endogenously Assembled RNA-Protein Complexes Using Affinity Purification Based on Streptavidin Aptamer S1. Int. J. Mol. Sci. 2015, 16, 22456-22472. https://doi.org/10.3390/ijms160922456
Dong Y, Yang J, Ye W, Wang Y, Ye C, Weng D, Gao H, Zhang F, Xu Z, Lei Y. Isolation of Endogenously Assembled RNA-Protein Complexes Using Affinity Purification Based on Streptavidin Aptamer S1. International Journal of Molecular Sciences. 2015; 16(9):22456-22472. https://doi.org/10.3390/ijms160922456
Chicago/Turabian StyleDong, Yangchao, Jing Yang, Wei Ye, Yuan Wang, Chuantao Ye, Daihui Weng, Huan Gao, Fanglin Zhang, Zhikai Xu, and Yingfeng Lei. 2015. "Isolation of Endogenously Assembled RNA-Protein Complexes Using Affinity Purification Based on Streptavidin Aptamer S1" International Journal of Molecular Sciences 16, no. 9: 22456-22472. https://doi.org/10.3390/ijms160922456
APA StyleDong, Y., Yang, J., Ye, W., Wang, Y., Ye, C., Weng, D., Gao, H., Zhang, F., Xu, Z., & Lei, Y. (2015). Isolation of Endogenously Assembled RNA-Protein Complexes Using Affinity Purification Based on Streptavidin Aptamer S1. International Journal of Molecular Sciences, 16(9), 22456-22472. https://doi.org/10.3390/ijms160922456