Insect Gallers and Their Plant Hosts: From Omics Data to Systems Biology
Abstract
:1. Introduction
2. The Galling Trait
2.1. Adaptive Significance of the Galling Trait
2.2. Development and Structure of Galls
3. The Molecular Mechanisms of Gall Induction
3.1. Effectors in Insect Galling
3.2. Phytohormones in Insect Galling
4. Plant Defence Responses against Galling Insects
4.1. Direct Defences
4.2. Indirect Defences
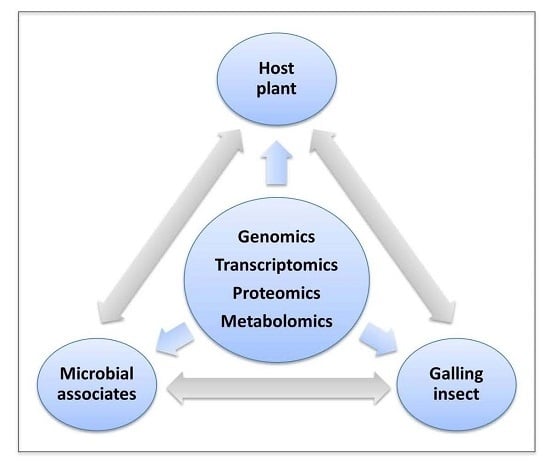
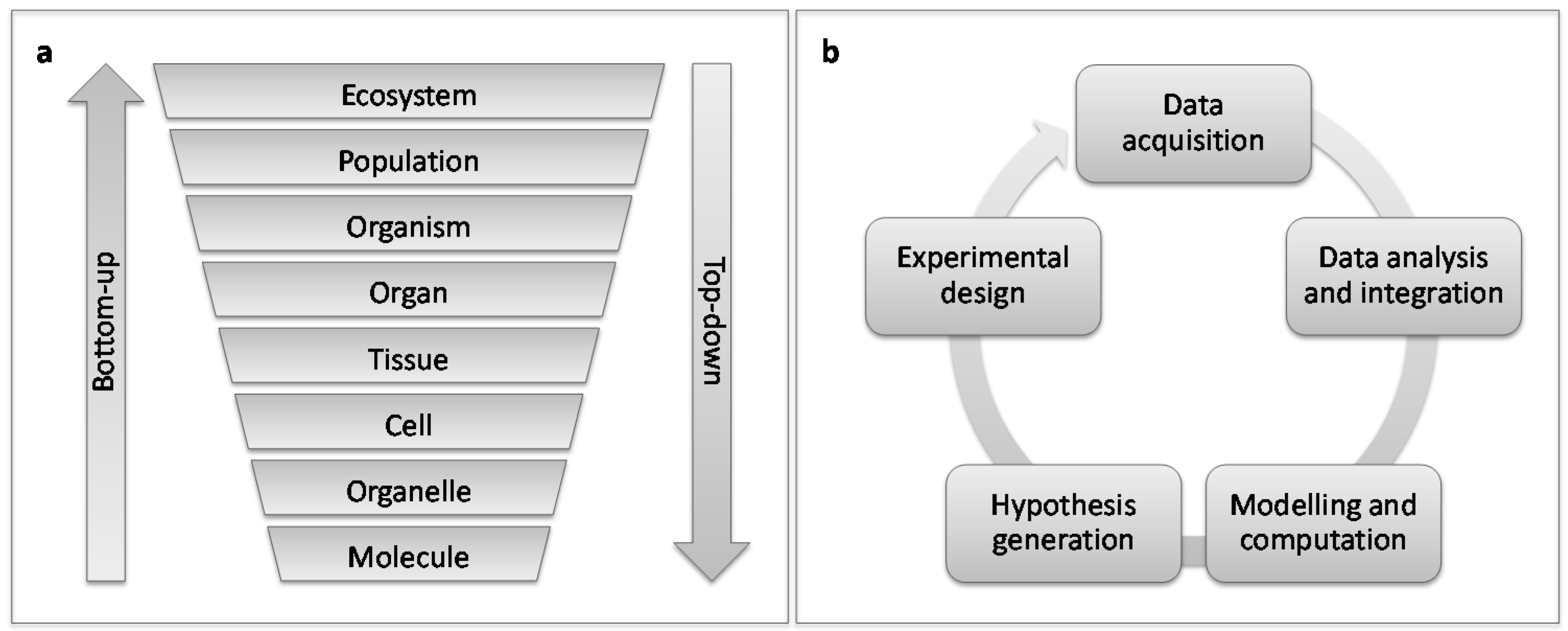
5. Host-Pest Interactions: Enter Systems Biology
6. Questions to Be Addressed Using Systems Biology Approaches for Plant-Galler Interactions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| R | Resistance |
| PTI | Pattern-triggered immunity |
| MAMPs | Microbe-associated molecular patterns |
| ETS | Effector-triggered susceptibility |
| ETI | Effector-triggered immunity |
| HAMPs | Herbivore-associated molecular patterns |
| EAMPs | Egg-associated molecular patterns |
| SSGP | Secreted salivary gland protein-encoding gene |
| miRNA | microRNA |
| IAA | Indole acetic acid |
| LC-MS-MS | Liquid chromatography coupled to tandem mass spectrometry |
References
- Stone, G.N.; Schönrogge, K. The adaptive significance of insect gall morphology. Trends Ecol. Evol. 2003, 18, 512–522. [Google Scholar] [CrossRef]
- Inbar, M.; Izhaki, I.; Koplovich, A.; Lupo, I.; Silanikove, N.; Glasser, T.; Gerchman, Y.; Perevolotsky, A.; Lev-Yadun, S. Why do many galls have conspicuous colors? A new hypothesis. Arthropod-Plant Interact. 2010, 4, 1–6. [Google Scholar] [CrossRef]
- Oliveira, D.C.; Isaias, R.M.S. Redifferentiation of leaflet tissues during midrib gall development in Copaifera langsdorffii (Fabaceae). S. Afr. J. Bot. 2010, 76, 239–248. [Google Scholar] [CrossRef]
- Tooker, J.F.; Rohr, J.R.; Abrahamson, W.G.; de Moraes, C.M. Gall insects can avoid and alter indirect plant defenses. New Phytol. 2008, 178, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Thrall, P.H.; Papa, J.; Xie, L.; Burdon, J.J. Playing on a Pathogen’s Weakness: Using Evolution to Guide Sustainable Plant Disease Control Strategies. Annu. Rev. Phytopathol. 2015, 53, 19–43. [Google Scholar] [CrossRef] [PubMed]
- Barah, P.; Bones, A.M. Multidimensional approaches for studying plant defence against insects: From ecology to omics and synthetic biology. J. Exp. Bot. 2015, 66, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Windram, O.; Penfold, C.A.; Denby, K.J. Network Modeling to Understand Plant Immunity. Annu. Rev. Phytopathol. 2014, 1–20. [Google Scholar]
- Schachat, S.R.; Labandeira, C.C. Evolution of a complex behavior: The origin and initial diversification of foliar galling by Permian insects. Sci. Nat. 2015, 102, 14. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Nieves-Aldrey, J.-L.; Buffington, M.L.; Liu, Z.; Liljeblad, J.; Nylander, J.A.A. Phylogeny, Evolution and Classification of Gall Wasps: The Plot Thickens. PLoS ONE 2015, 10, e0123301. [Google Scholar] [CrossRef] [PubMed]
- Giron, D.; Glevarec, G. Cytokinin-Induced Phenotypes in Plant-Insect Interactions: Learning from the Bacterial World. J. Chem. Ecol. 2014, 40, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Spíchal, L. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef]
- Kaiser, W.; Huguet, E.; Giron, D. Plant green-island phenotype induced by leaf-miners is mediated by bacterial symbionts. Proc. R. Soc. 2010. [Google Scholar] [CrossRef] [PubMed]
- Giron, D.; Kaiser, W.; Imbault, N. Cytokinin-mediated leaf manipulation by a leafminer caterpillar. Biol. Lett. 2007, 3, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.-Y.; Huang, W.-D.; Chou, H.-M.; Chen, C.-C.; Chen, P.-J.; Chang, Y.-T.; Yang, C.-M. Structural, biochemical, and physiological characterization of photosynthesis in leaf-derived cup-shaped galls on Litsea acuminata. BMC Plant Biol. 2015, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.K.; Moran, N.A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2014, 23, 1473–1496. [Google Scholar] [CrossRef] [PubMed]
- Giron, D.; Huguet, E.; Stone, G.N.; Body, M. Insect-induced effects on plants and possible effectors used by galling and leaf-mining insects to manipulate their host-plant. J. Insect Physiol. 2016, 84, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Dardeau, F.; Deprost, E.; Laurans, F.; Lainé, V.; Lieutier, F.; Sallé, A. Resistant poplar genotypes inhibit pseudogall formation by the wooly poplar aphid, phloeomyzus passerinii sign. Trees 2014, 28, 1007–1019. [Google Scholar] [CrossRef]
- Ferreira, B.G.; Teixeira, C.T.; Isaias, R.M.S. Efficiency of the Polyethylene-Glycol (PEG) Embedding Medium for Plant Histochemistry. J. Histochem. Cytochem. 2014, 62, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, B.G.; Isaias, R.M.S. Developmental stem anatomy and tissue redifferentiation induced by a galling Lepidoptera on Marcetia taxifolia (Melastomataceae). Botany 2013, 760, 752–760. [Google Scholar] [CrossRef]
- Vecchi, C.; Menezes, N.L.; Oliveira, D.C.; Ferreira, B.G.; Isaias, R.M.S. The redifferentiation of nutritive cells in galls induced by Lepidoptera on Tibouchina pulchra (Cham.) Cogn. reveals predefined patterns of plant development. Protoplasma 2013, 250, 1363–1368. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.A.; Body, M.; Warmund, M.R.; Schultz, J.C.; Appel, H.M. Morphometric analysis of young petiole galls on the narrow-leaf cottonwood, Populus angustifolia, by the sugarbeet root aphid, Pemphigus betae. Protoplasma 2016. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, B.G.; Carneiro, R.G.S.; Isaias, R.M.S. Multivesicular bodies differentiate exclusively in nutritive fast-dividing cells in Marcetia taxifolia galls. Protoplasma 2015, 252, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Oates, C.N.; Külheim, C.; Myburg, A.A.; Slippers, B.; Naidoo, S. The transcriptome and terpene profile of Eucalyptus grandis reveals mechanisms of defense against the insect pest, Leptocybe invasa. Plant Cell Physiol. 2015, 56, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.-Y.; Huang, W.-D.; Chou, H.-M.; Lin, K.-H.; Chen, C.-C.; Chen, P.-J.; Chang, Y.-T.; Yang, C.-M. Leaf-derived cecidomyiid galls are sinks in Machilus thunbergii (Lauraceae) leaves. Physiol. Plant. 2014, 152, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Nabity, P.D.; Haus, M.J.; Berenbaum, M.R.; DeLucia, E.H. Leaf-galling phylloxera on grapes reprograms host metabolism and morphology. Proc. Natl. Acad. Sci. USA 2013, 110, 16663–16668. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Neeraja, C.N.; Nair, S.; Bentur, J.S. Differential gene expression in gall midge susceptible rice genotypes revealed by suppressive subtraction hybridization (SSH) cDNA libraries and microarray analysis. Rice 2012, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Compson, Z.G.; Larson, K.C.; Zinkgraf, M.S.; Whitham, T.G. A genetic basis for the manipulation of sink-source relationships by the galling aphid Pemphigus batae. Oecologia 2011, 167, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, J.; Huang, L.; Zhu, L.; Liu, X.; Weng, N.; Reese, J.C.; Harris, M.; Stuart, J.J.; Chen, M.S. Gene expression of different wheat genotypes during attack by virulent and avirulent Hessian fly (Mayetiola destructor) larvae. J. Chem. Ecol. 2007, 33, 2171–2194. [Google Scholar] [CrossRef] [PubMed]
- Saltzmann, K.D.; Giovanini, M.P.; Zheng, C.; Williams, C.E. Virulent hessian fly larvae manipulate the free amino acid content of host wheat plants. J. Chem. Ecol. 2008, 34, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.Y.M.; Bedetti, C.S.; dos Santos Isaias, R.M. Detection and distribution of cell growth regulators and cellulose microfibrils during the development of Lopesia sp. galls on Lonchocarpus cultratus (Fabaceae). Botany 2015, 444, 435–444. [Google Scholar] [CrossRef]
- Tooker, J.F.; Helms, A.M. Phytohormone Dynamics Associated with Gall Insects, and their Potential Role in the Evolution of the Gall-Inducing Habit. J. Chem. Ecol. 2014, 40, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Favery, B.; Quentin, M.; Jaubert-Possamai, S.; Abad, P. Gall-forming root-knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J. Insect Physiol. 2015, 84, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Gheysen, G.; Mitchum, M.G. How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 2011, 14, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Formiga, A.T.; de Oliveira, D.C.; Ferreira, B.G.; Magalhães, T.A.; de Castro, A.C.; Fernandes, G.W.; dos Santos Isaias, R.M. The role of pectic composition of cell walls in the determination of the new shape-functional design in galls of Baccharis reticularia (Asteraceae). Protoplasma 2013, 250, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, R.G.S.; Oliveira, D.C.; Isaias, R.M.S. Developmental anatomy and immunocytochemistry reveal the neo-ontogenesis of the leaf tissues of Psidium myrtoides (Myrtaceae) towards the globoid galls of Nothotrioza myrtoidis (Triozidae). Plant Cell Rep. 2014, 33, 2093–2106. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.C.; Isaias, R.M.S.; Fernandes, G.W.; Ferreira, B.G.; Carneiro, R.G.S.; Fuzaro, L. Manipulation of host plant cells and tissues by gall-inducing insects and adaptive strategies used by different feeding guilds. J. Insect Physiol. 2016, 84, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, L.; Rivas, S. Catch me if you can: Bacterial effectors and plant targets. Trends Plant Sci. 2012, 17, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J. Insect effectors and gene-for-gene interactions with host plants. Curr. Opin. Insect Sci. 2015, 9, 56–61. [Google Scholar] [CrossRef]
- Villarroel, C.A.; Jonckheere, W.; Alba, J.M.; Glas, J.J.; Dermauw, W.; Haring, M.A.; van, T.; Schuurink, R.C.; Kant, M.R. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 2016, 86, 119–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshinaga, N.; Alborn, H.T.; Nakanishi, T.; Suckling, D.M.; Nishida, R.; Tumlinson, J.H.; Mori, N. Fatty acid-amino acid conjugates diversification in lepidopteran caterpillars. J. Chem. Ecol. 2010, 36, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Leclere, S.; Carroll, M.J.; Alborn, H.T.; Teal, P.E.A. Cowpea Chloroplastic ATP Synthase Is the Source of Multiple Plant Defense Elicitors during Insect Herbivory. Plant Physiol. 2007, 144, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Doss, R.P.; Oliver, J.E.; Proebsting, W.M.; Potter, S.W.; Kuy, S.; Clement, S.L.; Williamson, R.T.; Carney, J.R.; DeVilbiss, E.D. Bruchins: Insect-derived plant regulators that stimulate neoplasm formation. Proc. Natl. Acad. Sci. USA 2000, 97, 6218–6223. [Google Scholar] [CrossRef] [PubMed]
- Alborn, H.T.; Turlings, T.C.J.; Jones, T.H.; Stenhagen, G.; Loughrin, J.H.; Tumlinson, J.H. An Elicitor of Plant Volatiles from Beet Armyworm Oral Secretion. Science 1997, 276, 945–949. [Google Scholar] [CrossRef]
- Aggarwal, R.; Subramanyam, S.; Zhao, C.; Chen, M.S.; Harris, M.O.; Stuart, J.J. Avirulence effector discovery in a plant galling and plant parasitic arthropod, the Hessian fly (Mayetiola destructor). PLoS ONE 2014, 9, e100958. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, P.; Cock, P.J.A.; Bos, J. Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genom. 2016, 17, 172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Escalante, L.N.; Chen, H.; Benatti, T.R.; Qu, J.; Chellapilla, S.; Waterhouse, R.M.; Wheeler, D.; Andersson, M.N.; Bao, R.; et al. A Massive Expansion of Effector Genes Underlies Gall-Formation in the Wheat Pest Mayetiola destructor. Curr. Biol. 2015, 25, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, C.; Williams, C.E.; El Bouhssini, M.; Whitworth, R.J.; Richards, S.; Stuart, J.J.; Chen, M.-S. Deep sequencing and genome-wide analysis reveals the expansion of MicroRNA genes in the gall midge Mayetiola destructor. BMC Genom. 2013, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Kaloshian, I.; Walling, L.L. Hemipteran and dipteran pests: Effectors and plant host immune regulators. J. Integr. Plant Biol. 2016, 58, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Bos, J.I.B. Effector proteins that modulate plant–insect interactions. Curr. Opin. Plant Biol. 2011, 14, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Hilker, M.; Fatouros, N.E. Plant Responses to Insect Egg Deposition. Annu. Rev. Entomol. 2015, 60, 493–515. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.S.; Fellers, J.P.; Stuart, J.J.; Reese, J.C.; Liu, X. A group of related cDNAs encoding secreted proteins from Hessian fly [Mayetiola destructor (Say)] salivary glands. Insect Mol. Biol. 2004, 13, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhao, H.; Cheng, Y.; Scheffler, B.; Liu, X.; Liu, X.; Hulbert, S.; Stuart, J.J. Analysis of transcripts and proteins expressed in the salivary glands of Hessian fly (Mayetiola destructor) larvae. J. Insect Physiol 2008, 54, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.J.; Shukle, R.H.; Chen, M.-S.; Srivastava, S.; Subramanyam, S.; Schemerhorn, B.J.; Weintraub, P.G.; Abdel Moniem, H.E.M.; Flanders, K.L.; Buntin, G.D.; et al. Differential expression of candidate salivary effector proteins in field collections of Hessian fly, Mayetiola destructor. Insect Mol. Biol. 2015, 24, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; Carvunis, A.-R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Jiang, Z.; Peng, Y.-L.; Zhang, Z. Revealing Shared and Distinct Gene Network Organization in Arabidopsis Immune Responses by Integrative Analysis. Plant Physiol. 2015, 167, 1186–1203. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.; Percy, D.M.; Hefer, C.A.; Cronk, Q.C.B. The transcriptional landscape of insect galls: psyllid (Hemiptera) gall formation in Hawaiian Metrosideros polymorpha (Myrtaceae). BMC Genom. 2015, 16, 943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; de Bernonville, T.D.; Body, M.; Glevarec, G.; Reichelt, M.; Unsicker, S.; Brunea, M.; Renou, J.P.; Huguet, E.; Dubreuil, G.; et al. Leaf-mining by Phyllonorycter blancardella reprograms the host-leaf transcriptome to modulate phytohormones associated with nutrient mobilization and plant defense. J. Insect Physiol. 2016, 84, 114–127. [Google Scholar]
- Yamaguchi, H.; Tanaka, H.; Hasegawa, M.; Tokuda, M.; Asami, T.; Suzuki, Y. Phytohormones and willow gall induction by a gall-inducing sawfly. New Phytol. 2012, 196, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Sheth, B.P.; Thaker, V.S. Plant systems biology: Insights, advances and challenges. Planta 2014, 240, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Fürstenberg-Hägg, J.; Zagrobelny, M.; Bak, S. Plant Defense against Insect Herbivores. Int. J. Mol. Sci. 2013, 14, 10242–10297. [Google Scholar] [CrossRef] [PubMed]
- Unsicker, S.B.; Kunert, G.; Gershenzon, J. Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 2009, 12, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Büchel, K.; McDowell, E.; Nelson, W.; Descour, A.; Gershenzon, J.; Hilker, M.; Soderlund, C.; Gang, D.R.; Fenning, T.; Meiners, T. An elm EST database for identifying leaf beetle egg-induced defense genes. BMC Genom. 2012, 13, 242. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Constabel, C.P. Molecular analysis of poplar defense against herbivory: Comparison of wound- and insect elicitor-induced gene expression. New Phytol. 2006, 172, 617–635. [Google Scholar] [CrossRef] [PubMed]
- Shukle, R.H.; Subramanyam, S.; Saltzmann, K.A.; Williams, C.E. Ultrastructural changes in the midguts of Hessian fly larvae feeding on resistant wheat. J. Insect Physiol. 2010, 56, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Bode, R.F.; Halitschke, R. Herbivore damage-induced production and specific anti-digestive function of serine and cysteine protease inhibitors in tall goldenrod, Solidago altissima L. (Asteraceae). Planta 2013, 237, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, X.; Zhang, S.; Zhu, Y.C.; Whitworth, R.J.; Chen, M.S. Differential responses of wheat inhibitor-like genes to Hessian fly, Mayetiola destructor, attacks during compatible and incompatible interactions. J. Chem. Ecol. 2008, 34, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shukle, R.; Mittapalli, O.; Cheng, Y.; Reese, J.C.; Wang, H.; Hua, B.; Chen, M. The gut transcriptome of a gall midge, Mayetiola destructor. J. Insect Physiol. 2010, 56, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Nyman, T.; Julkunen-Tiitto, R. Manipulation of the phenolic chemistry of willows by gall-inducing sawflies. Proc. Natl. Acad. Sci. USA 2000, 97, 13184–13187. [Google Scholar] [CrossRef] [PubMed]
- Bruinsma, M.; Posthumus, M.A.; Mumm, R.; Mueller, M.J.; van Loon, J.J.A.; Dicke, M. Jasmonic acid-induced volatiles of Brassica oleracea attract parasitoids: Effects of time and dose, and comparison with induction by herbivores. J. Exp. Bot. 2009, 60, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Troncoso, C.; Becerra, J.; Perez, C.; Hernandez, V.; Martin, A.S.; Sanchez-olate, M.; Rios, D. Induction of defensive responses in Eucalyptus globulus (Labill) plants, against Ctenarytaina eucalypti (Maskell) (Hemiptera: Psyllidae). Am. J. Plant Sci. 2012, 2012, 589–595. [Google Scholar] [CrossRef]
- Damasceno, F.C.; Nicolli, K.P.; Caram̃o, E.B.; Soares, G.L.G.; Zini, C.A. Changes in the volatile organic profile of Schinus polygamus (Anacardiaceae) and Baccharis spicata (Asteraceae) induced by galling psyllids. J. Braz. Chem. Soc. 2010, 21, 556–563. [Google Scholar] [CrossRef]
- Tooker, J.F.; de Moraes, C.M. Feeding by Hessian fly [Mayetiola destructor (Say)] larvae does not induce plant indirect defences. Ecol. Entomol. 2007, 32, 153–161. [Google Scholar] [CrossRef]
- Rizzetto, L.; Cavalieri, D. Friend or foe: Using systems biology to elucidate interactions between fungi and their hosts. Trends Microbiol. 2011, 19, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Dix, A.; Vlaic, S.; Guthke, R.; Linde, J. Use of systems biology to decipher host e pathogen interaction networks and predict biomarkers. Clin. Microbiol. Infect. 2016, 22, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Graziosi, I.; Rieske, L.K. A plant pathogen causes extensive mortality in an invasive insect herbivore. Agric. For. Entomol. 2015, 17, 366–374. [Google Scholar] [CrossRef]
- Tozkar, C.Ö.; Kence, M.; Kence, A.; Huang, Q.; Evans, J.D. Metatranscriptomic analyses of honey bee colonies. Front. Genet. 2015, 6, 100. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.F.; Scharf, M.E. Metatranscriptome analysis reveals bacterial symbiont contributions to lower termite physiology and potential immune functions. BMC Genom. 2016, 17, 772. [Google Scholar] [CrossRef] [PubMed]
- Martinson, E.O.; Hackett, J.D.; Machado, C.A.; Arnold, A.E. Metatranscriptome Analysis of Fig Flowers Provides Insights into Potential Mechanisms for Mutualism Stability and Gall Induction. PLoS ONE 2015, 10, e0130745. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Polanski, K.; Rhodes, J.; Hill, C.; Zhang, P.; Jenkins, D.J.; Kiddle, S.J.; Jironkin, A.; Beynon, J.; Buchanan-wollaston, V.; Ott, S.; et al. Wigwams: Identifying gene modules co-regulated across multiple biological conditions. Bioinformatics 2014, 30, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Windram, O.P.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.J.; Cooke, E.J.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 2012, 24, 3530–3557. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Nambiar-Veetil, M.; Sangeetha, M.; Sowmiya Rani, K.S.; Aravinthakumar, V.; Selvakesavan, R.K.; Balasubramanian, A.; Venkatachalam, R.; Mary, A.; Jacob, J.; Krishna Kumar, N. Identification of insect-specific target genes for development of RNAi based control of the Eucalyptus gall pest Leptocybe invasa Fisher & La Salle (Hymenoptera: Eulophidae). BMC Proc. 2011, 5, 98. [Google Scholar]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oates, C.N.; Denby, K.J.; Myburg, A.A.; Slippers, B.; Naidoo, S. Insect Gallers and Their Plant Hosts: From Omics Data to Systems Biology. Int. J. Mol. Sci. 2016, 17, 1891. https://doi.org/10.3390/ijms17111891
Oates CN, Denby KJ, Myburg AA, Slippers B, Naidoo S. Insect Gallers and Their Plant Hosts: From Omics Data to Systems Biology. International Journal of Molecular Sciences. 2016; 17(11):1891. https://doi.org/10.3390/ijms17111891
Chicago/Turabian StyleOates, Caryn N., Katherine J. Denby, Alexander A. Myburg, Bernard Slippers, and Sanushka Naidoo. 2016. "Insect Gallers and Their Plant Hosts: From Omics Data to Systems Biology" International Journal of Molecular Sciences 17, no. 11: 1891. https://doi.org/10.3390/ijms17111891
APA StyleOates, C. N., Denby, K. J., Myburg, A. A., Slippers, B., & Naidoo, S. (2016). Insect Gallers and Their Plant Hosts: From Omics Data to Systems Biology. International Journal of Molecular Sciences, 17(11), 1891. https://doi.org/10.3390/ijms17111891