Assessment of Radiation Induced Therapeutic Effect and Cytotoxicity in Cancer Patients Based on Transcriptomic Profiling
Abstract
:1. Introduction
2. Results
2.1. Identification of Differentially Expressed Genes
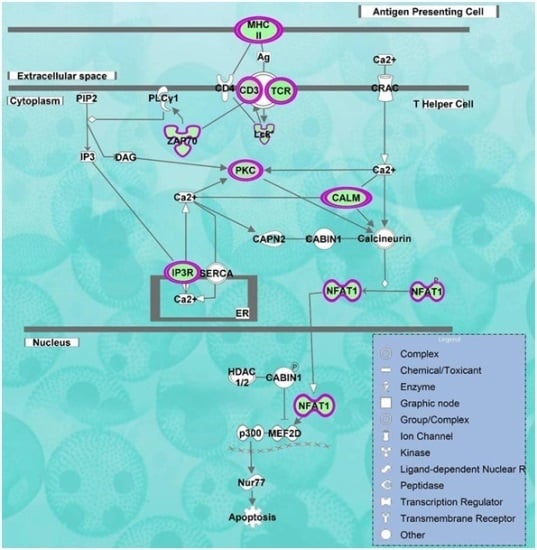
2.2. Pathways and Networks Underlying Immune Dysfunction
2.3. Toxicity Function Analysis
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Gene Expression Analysis
4.3. Functional and Pathway Analysis
4.4. Gene Set Enrichment Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GO | Gene ontology |
| RT | Radiation therapy |
| CA1 | carbonic anhydrase I |
| SNCA | a-synuclein |
| APCs | antigen-presenting cells |
| CRF | cancer related fatigue |
| MS4A1 | membrane-spanning 4-domains, subfamily A, member 1 |
References
- Jones, J.A.; Lutz, S.T.; Chow, E.; Johnstone, P.A. Palliative radiotherapy at the end of life: A critical review. CA Cancer J. Clin. 2014, 64, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.U.; Hunt, D.; McGowan, D.G.; Amin, M.B.; Chetner, M.P.; Bruner, D.W.; Leibenhaut, M.H.; Husain, S.M.; Rotman, M.; Souhami, L.; et al. Radiotherapy and short-term androgen deprivation for localized prostate cancer. N. Engl. J. Med. 2011, 365, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Thompson, I.; Thrasher, J.B.; Aus, G.; Burnett, A.L.; Canby-Hagino, E.D.; Cookson, M.S.; D’Amico, A.V.; Dmochowski, R.R.; Eton, D.T.; Forman, J.D.; et al. Guideline for the management of clinically localized prostate cancer: 2007 update. J. Urol. 2007, 177, 2106–2131. [Google Scholar] [PubMed]
- Rose, J.N.; Crook, J.M. The role of radiation therapy in the treatment of metastatic castrate-resistant prostate cancer. Ther. Adv. Urol. 2015, 7, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Saligan, L.N.; Hsiao, C.P.; Wang, D.; Wang, X.M.; St John, L.; Kaushal, A.; Citrin, D.; Barb, J.J.; Munson, P.J.; Dionne, R.A. Upregulation of α-synuclein during localized radiation therapy signals the association of cancer-related fatigue with the activation of inflammatory and neuroprotective pathways. Brain Behav. Immun. 2013, 27, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Chon, B.H.; Loeffler, J.S. The effect of nonmalignant systemic disease on tolerance to radiation therapy. Oncologist 2002, 7, 136–143. [Google Scholar] [CrossRef]
- Robertson, J.M.; Clarke, D.H.; Pevzner, M.M.; Matter, R.C. Breast conservation therapy. Severe breast fibrosis after radiation therapy in patients with collagen vascular disease. Cancer 1991, 68, 502–508. [Google Scholar] [CrossRef]
- Fleck, R.; McNeese, M.D.; Ellerbroek, N.A.; Hunter, T.A.; Holmes, F.A. Consequences of breast irradiation in patients with pre-existing collagen vascular diseases. Int. J. Radiat. Oncol. Biol. Phys. 1989, 17, 829–833. [Google Scholar] [CrossRef]
- Nieder, C.; Angelo, K.; Dalhaug, A.; Pawinski, A.; Haukland, E.; Norum, J. Palliative radiotherapy during the last month of life: Predictability for referring physicians and radiation oncologists. Oncol. Lett. 2015, 10, 3043–3049. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.; Bentzen, S.M.; Overgaard, J.; Overgaard, M. Relationship between the in vitro radiosensitivity of skin fibroblasts and the expression of subcutaneous fibrosis, telangiectasia, and skin erythema after radiotherapy. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 1996, 40, 101–109. [Google Scholar] [CrossRef]
- Crompton, N.E.; Miralbell, R.; Rutz, H.P.; Ersoy, F.; Sanal, O.; Wellmann, D.; Bieri, S.; Coucke, P.A.; Emery, G.C.; Shi, Y.Q.; et al. Altered apoptotic profiles in irradiated patients with increased toxicity. Int. J. Radiat. Oncol. Biol. Phys. 1999, 45, 707–714. [Google Scholar] [CrossRef]
- Barber, J.B.; Burrill, W.; Spreadborough, A.R.; Levine, E.; Warren, C.; Kiltie, A.E.; Roberts, S.A.; Scott, D. Relationship between in vitro chromosomal radiosensitivity of peripheral blood lymphocytes and the expression of normal tissue damage following radiotherapy for breast cancer. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2000, 55, 179–186. [Google Scholar] [CrossRef]
- Bokemeyer, M.; Ding, X.Q.; Goldbecker, A.; Raab, P.; Heeren, M.; Arvanitis, D.; Tillmann, H.L.; Lanfermann, H.; Weissenborn, K. Evidence for neuroinflammation and neuroprotection in hcv infection-associated encephalopathy. Gut 2011, 60, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Ronnback, L.; Hansson, E. On the potential role of glutamate transport in mental fatigue. J. Neuroinflamm. 2004, 1, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.M.; Guha, C.; Hodge, J.W.; Jaffee, E. Introduction: Immunobiology of radiotherapy: New paradigms. Radiat. Res. 2014, 182, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Srikrishna, G.; Freeze, H.H. Endogenous damage-associated molecular pattern molecules at the crossroads of inflammation and cancer. Neoplasia 2009, 11, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, J.M.; Communi, D. Modulation of inflammation by extracellular nucleotides. J. Investig. Dermatol. 2006, 126, 943–944. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Evans, J.E.; Rock, K.L. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature 2003, 425, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Farkas, A.M.; Kilgore, T.M.; Lotze, M.T. Detecting DNA: Getting and begetting cancer. Curr. Opin. Investig. Drugs 2007, 8, 981–986. [Google Scholar] [PubMed]
- Rotow, J.; Gameiro, S.R.; Madan, R.A.; Gulley, J.L.; Schlom, J.; Hodge, J.W. Vaccines as monotherapy and in combination therapy for prostate cancer. Clin. Transl. Sci. 2010, 3, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Obeid, M.; Ortiz, C.; Criollo, A.; Mignot, G.; Maiuri, M.C.; Ullrich, E.; Saulnier, P.; et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007, 13, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Hannani, D.; Poirier-Colame, V.; Ladoire, S.; Locher, C.; Sistigu, A.; Prada, N.; Adjemian, S.; Catani, J.P.; Freudenberg, M.; et al. Defective immunogenic cell death of hmgb1-deficient tumors: Compensatory therapy with tlr4 agonists. Cell Death Differ. 2014, 21, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Hofman, M.; Ryan, J.L.; Figueroa-Moseley, C.D.; Jean-Pierre, P.; Morrow, G.R. Cancer-related fatigue: The scale of the problem. Oncologist 2007, 12, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Saligan, L.N.; Olson, K.; Filler, K.; Larkin, D.; Cramp, F.; Yennurajalingam, S.; Escalante, C.P.; del Giglio, A.; Kober, K.M.; Kamath, J.; et al. The biology of cancer-related fatigue: A review of the literature. Support. Care Cancer 2015, 23, 2461–2478. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S. Pathophysiology of cancer-related fatigue. Clin. J. Oncol. Nurs. 2008, 12, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.M.; Zhang, F.; Zhou, H.; Kam, W.; Wilson, B.; Hong, J.S. Neuroinflammation and α-synuclein dysfunction potentiate each other, driving chronic progression of neurodegeneration in a mouse model of parkinson’s disease. Environ. Health Perspect. 2011, 119, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Sly, W.S.; Hu, P.Y. Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu. Rev. Biochem. 1995, 64, 375–401. [Google Scholar] [CrossRef] [PubMed]
- Tedder, T.F.; Streuli, M.; Schlossman, S.F.; Saito, H. Isolation and structure of a cDNA encoding the B1 (CD20) cell-surface antigen of human b lymphocytes. Proc. Natl. Acad. Sci. USA 1988, 85, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, T.; Tomita, A.; Sugimoto, K.; Shimada, K.; Iriyama, C.; Hirose, T.; Shirahata-Adachi, M.; Suzuki, Y.; Mizuno, H.; Kiyoi, H.; et al. De novo diffuse large B-cell lymphoma with a CD20 immunohistochemistry-positive and flow cytometry-negative phenotype: Molecular mechanisms and correlation with rituximab sensitivity. Cancer Sci. 2014, 105, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.-P.; Reddy, S.Y.; Chen, M.-K.; Saligan, L.N. Genomic profile of fatigued men receiving localized radiation therapy. Biol. Res. Nurs. 2015. [Google Scholar] [CrossRef] [PubMed]
- Dressman, H.K.; Muramoto, G.G.; Chao, N.J.; Meadows, S.; Marshall, D.; Ginsburg, G.S.; Nevins, J.R.; Chute, J.P. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007, 4. [Google Scholar] [CrossRef] [PubMed]
- Schubert, C.; Hong, S.; Natarajan, L.; Mills, P.J.; Dimsdale, J.E. The association between fatigue and inflammatory marker levels in cancer patients: A quantitative review. Brain Behav. Immun. 2007, 21, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.M.; Yu, Z.X.; Ferrans, V.J.; Lowenstein, R.A.; Finkel, T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc. Natl. Acad. Sci. USA 1996, 93, 11848–11852. [Google Scholar] [CrossRef] [PubMed]
- Kalimuthu, S.; Se-Kwon, K. Cell survival and apoptosis signaling as therapeutic target for cancer: Marine bioactive compounds. Int. J. Mol. Sci. 2013, 14, 2334–2354. [Google Scholar] [CrossRef] [PubMed]
- Chinnaiyan, A.M. The apoptosome: Heart and soul of the cell death machine. Neoplasia 1999, 1, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.M.; Adrain, C.; Duriez, P.J.; Creagh, E.M.; Martin, S.J. Analysis of the composition, assembly kinetics and activity of native apaf-1 apoptosomes. EMBO J. 2004, 23, 2134–2145. [Google Scholar] [CrossRef] [PubMed]
- Maor, Y.; Malnick, S. Liver injury induced by anticancer chemotherapy and radiation therapy. Int. J. Hepatol. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W.; Pollock, R.E.; Weichselbaum, R.R.; Bast, R.C., Jr.; Gansler, T.S.; Holland, J.F.; Frei, E. Holland-Frei Cancer Medicine, 6th ed.; BC Decker: Hamilton, ON, USA, 2003. [Google Scholar]
- Darby, S.C.; Ewertz, M.; McGale, P.; Bennet, A.M.; Blom-Goldman, U.; Bronnum, D.; Correa, C.; Cutter, D.; Gagliardi, G.; Gigante, B.; et al. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N. Engl. J. Med. 2013, 368, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Pistevou-Gompaki, K.; Hatzitolios, A.; Eleftheriadis, N.; Boultoukas, E.; Ntaios, G.; Andronikidis, I.; Tzitzikas, I. Evaluation of cardiotoxicity five years after 2D planned, non-simulated, radiation therapy for left breast cancer. Ther. Clin. Risk Manag. 2008, 4, 1359–1362. [Google Scholar] [PubMed]
- Yeh, E.T.H.; Tong, A.T.; Lenihan, D.J.; Yusuf, S.W.; Swafford, J.; Champion, C.; Durand, J.-B.; Gibbs, H.; Zafarmand, A.A.; Ewer, M.S. Cardiovascular complications of cancer therapy: Diagnosis, pathogenesis, and management. Circulation 2004, 109, 3122–3131. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, S.W.; Sami, S.; Daher, I.N. Radiation-induced heart disease: A clinical update. Cardiol. Res. Pract. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, M.P.; Pollok, M.; Fries, J.; Baldamus, C.A. Radiation nephropathy after radiotherapy in metastatic medullary thyroid carcinoma. Nephrol. Dial. Transplant. 2001, 16, 1082–1083. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, B.D.; Soiffer, R.J.; Magee, C.C. Renal failure associated with cancer and its treatment: An update. J. Am. Soc. Nephrol. 2005, 16, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Savitsky, K.; Bar-Shira, A.; Gilad, S.; Rotman, G.; Ziv, Y.; Vanagaite, L.; Tagle, D.A.; Smith, S.; Uziel, T.; Sfez, S.; et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995, 268, 1749–1753. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.S.; Maser, R.S.; Stankovic, T.; Bressan, D.A.; Kaplan, M.I.; Jaspers, N.G.; Raams, A.; Byrd, P.J.; Petrini, J.H.; Taylor, A.M. The DNA double-strand break repair gene hmre11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 1999, 99, 577–587. [Google Scholar] [CrossRef]
- Varon, R.; Vissinga, C.; Platzer, M.; Cerosaletti, K.M.; Chrzanowska, K.H.; Saar, K.; Beckmann, G.; Seemanova, E.; Cooper, P.R.; Nowak, N.J.; et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in nijmegen breakage syndrome. Cell 1998, 93, 467–476. [Google Scholar] [CrossRef]
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001, 105, 177–186. [Google Scholar] [CrossRef]
- West, C.M.; Elyan, S.A.; Berry, P.; Cowan, R.; Scott, D. A comparison of the radiosensitivity of lymphocytes from normal donors, cancer patients, individuals with ataxia-telangiectasia (A-T) and A-T heterozygotes. Int. J. Radiat. Biol. 1995, 68, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.J.; Schiff, P.B.; Hanks, G.E.; Brenner, D.J.; Russo, J.; Chen, J.; Sawant, S.G.; Pandita, T.K. A preliminary report: Frequency of A-T heterozygotes among prostate cancer patients with severe late responses to radiation therapy. Cancer J. Sci. Am. 1998, 4, 385–389. [Google Scholar] [PubMed]
- Ramsay, J.; Birrell, G.; Lavin, M. Testing for mutations of the ataxia telangiectasia gene in radiosensitive breast cancer patients. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 1998, 47, 125–128. [Google Scholar] [CrossRef]
- Oppitz, U.; Bernthaler, U.; Schindler, D.; Sobeck, A.; Hoehn, H.; Platzer, M.; Rosenthal, A.; Flentje, M. Sequence analysis of the atm gene in 20 patients with RTOG grade 3 or 4 acute and/or late tissue radiation side effects. Int. J. Radiat. Oncol. Biol. Phys. 1999, 44, 981–988. [Google Scholar] [CrossRef]
- Da Costa Miranda, V.; Trufelli, D.C.; Santos, J.; Campos, M.P.; Nobuo, M.; da Costa Miranda, M.; Schlinder, F.; Riechelmann, R.; del Giglio, A. Effectiveness of guarana (paullinia cupana) for postradiation fatigue and depression: Results of a pilot double-blind randomized study. J. Altern. Complement. Med. 2009, 15, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.P.; Hassan, B.J.; Riechelmann, R.; Del Giglio, A. Cancer-related fatigue: A review. Rev. Assoc. Med. Bras. 2011, 57, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Vijayalaxmi; Reiter, R.J.; Tan, D.X.; Herman, T.S.; Thomas, C.R., Jr. Melatonin as a radioprotective agent: A review. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Shirazi, A.; Ghobadi, G.; Ghazi-Khansari, M. A radiobiological review on melatonin: A novel radioprotector. J. Radiat. Res. 2007, 48, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Kucuktulu, E. Protective effect of melatonin against radiation induced nephrotoxicity in rats. Asian Pac. J. Cancer Prev. 2012, 13, 4101–4105. [Google Scholar] [CrossRef] [PubMed]
- Jagetia, G.C. Radioprotection and radiosensitization by curcumin. Adv. Exp. Med. Biol. 2007, 595, 301–320. [Google Scholar] [PubMed]
- Ben-Josef, E.; Han, S.; Tobi, M.; Vargas, B.J.; Stamos, B.; Kelly, L.; Biggar, S.; Kaplan, I. Intrarectal application of amifostine for the prevention of radiation-induced rectal injury. Semin. Radiat. Oncol. 2002, 12, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Zabbarova, I.; Kanai, A. Targeted delivery of radioprotective agents to mitochondria. Mol. Interv. 2008, 8, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Simone, N.L.; Menard, C.; Soule, B.P.; Albert, P.S.; Guion, P.; Smith, S.; Godette, D.; Crouse, N.S.; Sciuto, L.C.; Cooley-Zgela, T.; et al. Intrarectal amifostine during external beam radiation therapy for prostate cancer produces significant improvements in quality of life measured by epic score. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Amundson, S.A. Functional genomics and a new era in radiation biology and oncology. BioScience 2008, 58, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Dellas, K. Does radiotherapy have curative potential in metastatic patients? The concept of local therapy in oligometastatic breast cancer. Breast Care 2011, 6, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Mirza, Z.; Schulten, H.J.; Farsi, H.M.; Al-Maghrabi, J.A.; Gari, M.A.; Chaudhary, A.G.; Abuzenadah, A.M.; Al-Qahtani, M.H.; Karim, S. Molecular interaction of a kinase inhibitor midostaurin with anticancer drug targets, S100A8 and EGFR: Transcriptional profiling and molecular docking study for kidney cancer therapeutics. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Merdad, A.; Karim, S.; Schulten, H.J.; Jayapal, M.; Dallol, A.; Buhmeida, A.; Al-Thubaity, F.; Gari, I.M.; Chaudhary, A.G.; Abuzenadah, A.M.; et al. Transcriptomics profiling study of breast cancer from kingdom of Saudi Arabia revealed altered expression of adiponectin and fatty acid binding protein4: Is lipid metabolism associated with breast cancer? BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]




| S. No | Gene Symbol | Gene Title | Chromosome Location | Fold-Change | p-Value |
|---|---|---|---|---|---|
| 1 | SNCA | synuclein, alpha (non A4 component of amyloid precursor) | chr4q21 | 3.50 | 2.70 × 10−6 |
| 2 | CA1 | carbonic anhydrase I | chr8q21.2 | 3.30 | 9.12 × 10−7 |
| 3 | XK | X-linked Kx blood group | chrXp21.1 | 2.86 | 0.000510 |
| 4 | GYPB | glycophorin B (MNS blood group) | chr4q31.21 | 2.83 | 1.25 × 10−6 |
| 5 | HEMGN | hemogen | chr9q22.33 | 2.82 | 4.06 × 10−5 |
| 6 | GYPA | glycophorin A (MNS blood group) | chr4q31.21 | 2.67 | 1.13 × 10−5 |
| 7 | BPGM | 2,3-bisphosphoglycerate mutase | chr7q33 | 2.47 | 0.002613 |
| 8 | FAM46C | family with sequence similarity 46, member C | chr1p12 | 2.40 | 0.002343 |
| 9 | ABCC13 | ATP-binding cassette, sub-family C (CFTR/MRP), member 13, pseudogene | chr21q11.2 | 2.35 | 1.87 × 10−5 |
| 10 | FECH | ferrochelatase | 18q21.31 | 2.33 | 0.001334 |
| 11 | ISCA1 | iron-sulfur cluster assembly 1 | chr9q21.33 | 2.32 | 1.7 × 10−6 |
| 12 | CCDC176 | coiled-coil domain containing 176 | chr14q24.3 | 2.29 | 0.000566 |
| 13 | AHSP | alpha hemoglobin stabilizing protein | chr16p11.2 | 2.29 | 7.18 × 10−7 |
| 14 | YOD1 | YOD1 deubiquitinase | chr1q32.2 | 2.25 | 0.000749 |
| 15 | NUDT4//NUDT4P1 | nudix (nucleoside diphosphate linked moiety X)-type motif 4 | chr12q21//chr1p12-p13 | 2.19 | 6.08 × 10−5 |
| 16 | RHD | Rh blood group, D antigen | 1p36.11 | 2.18 | 1.02 × 10−7 |
| 17 | FGFR1OP2 | FGFR1 oncogene partner 2 | chr12p11.23 | 2.04 | 0.000354 |
| 18 | TSPO2 | translocator protein 2 | 6p21.1 | 2.03 | 6.71 × 10−13 |
| 19 | ITLN1 | intelectin 1 (galactofuranose binding) | 1q23.3 | 2.02 | 6.64 × 10−6 |
| 20 | KRT1 | keratin 1 | 12q13.13 | 2.01 | 0.002745 |
| Ingenuity Canonical Pathways | -log (p-Value) | z-Score | Molecules |
|---|---|---|---|
| Calcium-induced T Lymphocyte Apoptosis | 10.8 | −3.317 | CD247, CD3G, LCK, PRKCQ, CAMK4, TRGV9, ZAP70, NFATC2, HLA-DOB, PRKCH, ITPR1, CD3D, PRKCA |
| Role of NFAT in Regulation of the Immune Response | 10.5 | −3.771 | CD247, BLNK, FYN, CAMK4, PRKCQ, NFATC3,T RGV9, ITPR1, CD3D, CD3G, LCK, RRAS2, LAT, ZAP70, HLA-DOB, RCAN3, NFATC2, IKBKAP, ATM,ITK |
| iCOS-iCOSL Signaling in T Helper Cells | 10.3 | −3.000 | CD247, CAMK4, PRKCQ, NFATC3, TRGV9, ITPR1, CD3D, CD3G, LCK, ZAP70, LAT, NFATC2, HLA-DOB, PLEKHA1, ATM,ITK |
| CD28 Signaling in T Helper Cells | 9.61 | −3.317 | CD247, FYN, CAMK4, PRKCQ, NFATC3, TRGV9, ITPR1, CD3D, CD3G, LCK, ZAP70, LAT, NFATC2, HLA-DOB, ATM,ITK |
| PKCθ Signaling in T Lymphocytes | 8.64 | −2.324 | CD247,F YN, PRKCQ, NFATC3, TRGV9, MAP3K4, CD3D, CD3G, LCK,RRAS2, ZAP70, LAT, NFATC2, HLA-DOB, ATM |
| Phospholipase C Signaling | 8.01 | −3.606 | CD247,BLNK, PEBP1, FYN, CAMK4, PRKCQ, NFATC3, TRGV9, ITPR1, CD3D, RHOH, CD3G, LCK, RRAS2, LAT, ZAP70, NFATC2, PRKCH, PRKCA, ITK |
| Tec Kinase Signaling | 5.18 | −3.606 | FYN, PRKCQ, TRGV9, RHOH, STAT4, BLK, LCK, TXK, TNFRSF25, PRKCH, ITK, PRKCA, ATM |
| EIF2 Signaling | 4.62 | −2.828 | RPL22, RPS18, RPS4X, RPL10A, RPL14, RRAS2, RPS20, RPL5, RPL36, RPL18, EIF3L, RPS24, ATM |
| B Cell Receptor Signaling | 4.02 | −1.897 | BLNK, PAX5, ETS1, EBF1, CAMK4, PRKCQ, RRAS2, FOXO1, NFATC3, NFATC2, MAP3K4, ATM |
| PI3K Signaling in B Lymphocytes | 3.88 | −2.828 | CD81, BLNK, BLK, FYN, CAMK4, RRAS2, NFATC3, NFATC2, PLEKHA1, ITPR1 |
| fMLP Signaling in Neutrophils | 3.66 | −3.000 | CAMK4, PRKCQ, RRAS2, NFATC3, NFATC2, PRKCH, ITPR1, PRKCA, ATM |
| Apoptosis Signaling | 1.56 | 2.236 | PRKCQ, RRAS2, BIRC3, PRKCA, BCL2 |
| Cytotoxic T Lymphocyte-mediated Apoptosis of Target Cells | 3.84 | 1.342 | CD247, CD3G, TRGV9, CD3D, BCL2 |
| Functional Category | Function Annotations | p-Value | Molecules |
|---|---|---|---|
| Cardiotoxicity | |||
| Cardiac Proliferation | proliferation of cardiomyocytes | 1.07 × 10−1 | FOXP1, NOG |
| Cardiac Arteriopathy | coronary artery disease | 5.09 × 10−1 | ABCG1, CD47, DOCK9, MARCH6, PDE7A, PRKCH |
| Cardiac Necrosis/Cell Death | apoptosis of cardiomyocytes and ventricular myocytes | 5.36 × 10−1 | BNIP3, NOG |
| Heart Failure | chronic heart failure | 4.73 × 10−1 | CA1 |
| Cardiac Infarction | myocardial infarction | 1.00 × 10−1 | CD47, MIAT |
| Hepatotoxicity | |||
| Liver Damage | low and high grade chronic hepatitis C, chronic hepatitis C, hepatotoxicity | 1.92 × 10−2 | CCR7, IMPDH2, RASGRP1 |
| Liver Hyperplasia/Hyper-proliferation | inflammatory hepatocellular adenoma; hepatocellular carcinoma; growth of hepatocellular carcinoma; liver cancer | 7.47 × 10−2 | IL6ST, MYC, + 113 genes |
| Liver Inflammation/Hepatitis | inflammation of liver; steatohepatitis; chronic hepatitis C | 3.26 × 10−1 | CCR7, IMPDH2, LPIN1, PDE7A |
| Liver Steatosis | hepatic steatosis; steatohepatitis; nonalcoholic steatohepatitis | 1.85 × 10−1 | LPIN1, PDE7A, RORA |
| Liver Fibrosis | fibrosis of liver; activation, migration and proliferation of hepatic stellate cells | 1.39 × 10−1 | IL6ST, RORA, CCR7 |
| Liver Necrosis/Cell Death | cell death of liver cells; apoptosis of hepatocytes | 3.91 × 10−1 | BCL2, MYC |
| Liver Proliferation | proliferation of liver cells; proliferation of hepatocytes; proliferation of hepatic stellate cells | 2.24 × 10−1 | IL6ST, LY9, MYC |
| Nephrotoxicity | |||
| Renal Necrosis/Cell Death | apoptosis of kidney cell lines; apoptosis of podocytes; cell death of kidney cell lines; cell viability of kidney cell lines | 5.51 × 10−2 | AQP3,AAK1,BCL2, BIRC3, BNIP3, DDX17, FOXO1, ITPR1, MYC, PRKCA, SNCA, TNFRSF25 |
| Nephrosis | nephrosis; minimal change nephrotic syndrome; autosomal recessive steroid-resistant nephrotic syndrome; steroid dependent nephrotic syndrome | 2.43 × 10−1 | IMPDH2, MS4A1 |
| Renal Nephritis | IgA nephropathy; membranous glomerulonephritis; lupus nephritis | 1.13 × 10−1 | IMPDH2, MS4A1 |
| Renal Proliferation | proliferation of mesangial cells; proliferation of kidney cell lines | 3.96 × 10−1 | CCR7, HSP90AB1, KMT2A, SFPQ |
| Kidney Failure | end stage renal disease | 4.63 × 10−1 | IMPDH2, PDE7A |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karim, S.; Mirza, Z.; Chaudhary, A.G.; Abuzenadah, A.M.; Gari, M.; Al-Qahtani, M.H. Assessment of Radiation Induced Therapeutic Effect and Cytotoxicity in Cancer Patients Based on Transcriptomic Profiling. Int. J. Mol. Sci. 2016, 17, 250. https://doi.org/10.3390/ijms17020250
Karim S, Mirza Z, Chaudhary AG, Abuzenadah AM, Gari M, Al-Qahtani MH. Assessment of Radiation Induced Therapeutic Effect and Cytotoxicity in Cancer Patients Based on Transcriptomic Profiling. International Journal of Molecular Sciences. 2016; 17(2):250. https://doi.org/10.3390/ijms17020250
Chicago/Turabian StyleKarim, Sajjad, Zeenat Mirza, Adeel G. Chaudhary, Adel M. Abuzenadah, Mamdooh Gari, and Mohammed H. Al-Qahtani. 2016. "Assessment of Radiation Induced Therapeutic Effect and Cytotoxicity in Cancer Patients Based on Transcriptomic Profiling" International Journal of Molecular Sciences 17, no. 2: 250. https://doi.org/10.3390/ijms17020250
APA StyleKarim, S., Mirza, Z., Chaudhary, A. G., Abuzenadah, A. M., Gari, M., & Al-Qahtani, M. H. (2016). Assessment of Radiation Induced Therapeutic Effect and Cytotoxicity in Cancer Patients Based on Transcriptomic Profiling. International Journal of Molecular Sciences, 17(2), 250. https://doi.org/10.3390/ijms17020250