Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer
Abstract
:1. Introduction
2. MicroRNAs: Biogenesis, Biological Role and Involvement in Drug Resistance
3. MicroRNAs as Regulators of Drug Resistance Pathways in GC
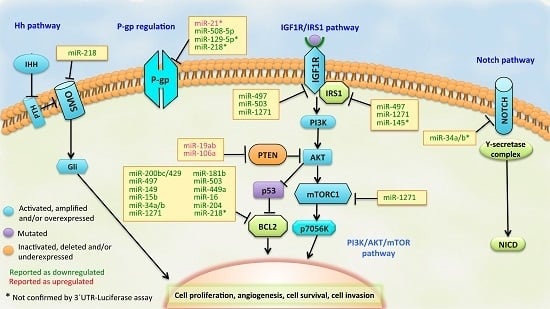
3.1. BCL2 Pathway
3.2. PI3K/PTEN/AKT Pathway
3.3. IGF1R/IRS1 Pathway
3.4. ABCB1 (MDR1/P-gp) Regulation
3.5. Other Signaling Pathways of Drug Resistance in GC
4. miRNA Signatures in Drug-Resistant GC
5. Concluding Remarks
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCB1 | ATP-binding cassette, sub-family B, member 1 |
| IGF1R | insulin-like growth factor 1 receptor |
| IRS1 | insulin receptor substrate 1 |
| ZNRD1 | zinc ribbon domain-containing |
| PTEN | phosphatase and tension homolog |
| BCL2 | B-cell lymphoma 2 |
| XIAP | X-linked inhibitor of apoptosis protein |
| RUNX3 | runt-related transcription factor 3 |
| SMO | Smoothened |
| MAPT | microtubule associated protein tau |
| HMGA2 | high-mobility group AT-hook 2 |
| IRF1 | interferon regulator factor 1 |
| VCR | vincristine |
| CDDP | cisplatin |
| ADR | adriamycin |
| VP-16 | etoposide |
| 5-Fu | 5-fluoruracil |
| DOX | doxorubicin |
| DTX | docetaxel |
| GEM | GEM |
| PTX | paclitaxel |
| L-OHP | oxaliplatin |
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA. Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Catalano, V.; Labianca, R.; Beretta, G.D.; Gatta, G.; de Braud, F.; Van Cutsem, E. Gastric cancer. Crit. Rev. Oncol. Hematol. 2009, 71, 127–164. [Google Scholar] [CrossRef] [PubMed]
- Lippert, T.H.; Ruoff, H.-J.; Volm, M. Intrinsic and acquired drug resistance in malignant tumors. The main reason for therapeutic failure. Arzneimittelforschung 2008, 58, 261–264. [Google Scholar] [PubMed]
- Rabik, C.A.; Dolan, M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 2007, 33, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.W.; Ruefli, A.A.; Lowe, S.W. Apoptosis: A link between cancer genetics and chemotherapy. Cell 2002, 108, 153–164. [Google Scholar] [CrossRef]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Fojo, T. Multiple paths to a drug resistance phenotype: mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug Resist. Updates 2007, 10, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Glasspool, R.M.; Teodoridis, J.M.; Brown, R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br. J. Cancer 2006, 94, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013, 774, 1–20. [Google Scholar] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M.; Calin, G.A. miRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; Kim, V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Filippov, V.; Solovyev, V.; Filippova, M.; Gill, S.S. A novel type of RNase III family proteins in eukaryotes. Gene 2000, 245, 213–221. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.-W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.-W.; MacRae, I.J. The molecular machines that mediate microRNA maturation. J. Cell. Mol. Med. 2009, 13, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.O.; Lai, E.C. Mirtrons: microRNA biogenesis via splicing. Biochimie 2011, 93, 1897–1904. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Chung, W.-J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Mol. Cell 2007, 28, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate miR-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. PNAS 2010, 107, 15163–15168. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.S. OncomiRs: The discovery and progress of microRNAs in cancers. Mol. Cancer 2007, 6. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Wang, J.; Chen, X.; Liu, L. Role of microRNA in anticancer drug resistance. Int. J. Cancer 2010, 126, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.-M. Small Interfering RNA in Drug Metabolism and Transport. Curr. Drug Metab. 2007, 8, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Leonard, G.D.; Fojo, T.; Bates, S.E. The role of ABC transporters in clinical practice. Oncologist 2003, 8, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xu, H.; Zhu, D.; Zhi, H.; Wang, T.; Wang, J.; Jiang, B.; Shu, Y.; Liu, P. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother. Pharmacol. 2012, 69, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Shan, X.; Wang, T.; Shu, Y.; Liu, P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int. J. Cancer 2010, 127, 2520–2529. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhu, D.; Lu, S.; Wang, T.; Wang, J.; Jiang, B.; Shu, Y.; Liu, P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med. Oncol. 2012, 29, 384–391. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Qian, J.; Shi, Q.; Zhu, J. miR-497 regulates cisplatin resistance of human gastric cancer cell line by targeting IGF1R, IRS1 and BCL2. J. Med. Oncol. 2015, 2, 1–7. [Google Scholar]
- Wang, T.; Ge, G.; Ding, Y.; Zhou, X.; Huang, Z.; Zhu, W.; Shu, Y.; Liu, P. miR-503 regulates cisplatin resistance of human gastric cancer cell lines by targeting IGF1R and BCL2. Chin. Med. J. (Engl.) 2014, 127, 2357–2362. [Google Scholar] [PubMed]
- Shu, Y. AB50. miR-503 modulates drug resistance of human gastric cancer cell lines by targeting BCL2. Transl. Gastrointest. Cancer 2013, 2. [Google Scholar] [CrossRef]
- Zhuang, M.; Shi, Q.; Zhang, X.; Ding, Y.; Shan, L.; Shan, X.; Qian, J.; Zhou, X.; Huang, Z.; Zhu, W.; et al. Involvement of miR-143 in cisplatin resistance of gastric cancer cells via targeting IGF1R and BCL2. Tumour Biol. 2015, 36, 2737–2745. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Iio, A.; Nakagawa, Y.; Naoe, T.; Tanigawa, N.; Akao, Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 2009, 77, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fang, Y.; Cao, Y.; Qin, R.; Chen, Q. miR-449a regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig. Dis. Sci. 2014, 59, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 2008, 123, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Hao, X.; Meng, Y.; Zhang, M.; Desano, J.; Fan, D.; Xu, L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Sacconi, A.; Biagioni, F.; Canu, V.; Mori, F.; Di Benedetto, A.; Lorenzon, L.; Ercolani, C.; Di Agostino, S.; Cambria, A.M.; Germoni, S.; et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, T.; Zhang, B.; Li, H.; Wu, Q.; Yang, L.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem. Biophys. Res. Commun. 2013, 434, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Shen, H.; Li, H.; Cao, Y.; Qin, R.; Long, L.; Zhu, X.; Xie, C.; Xu, W. miR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim. Biophys. Sin. 2013, 45, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, Q.; Cai, X. MicroRNA-106a induces multidrug resistance in gastric cancer by targeting RUNX3. FEBS Lett. 2013, 587, 3069–3075. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-M.; Huang, C.; Li, X.-F.; Yu, M.-Z.; He, Y.; Li, J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology 2013, 306, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Eto, K.; Iwatsuki, M.; Watanabe, M.; Ida, S.; Ishimoto, T.; Iwagami, S.; Baba, Y.; Sakamoto, Y.; Miyamoto, Y.; Yoshida, N.; et al. The microRNA-21/PTEN pathway regulates the sensitivity of HER2-positive gastric cancer cells to trastuzumab. Ann. Surg. Oncol. 2014, 21, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Liu, Y.; Wang, H. Antagonism of miRNA-21 sensitizes human gastric cancer cells to paclitaxel. Cell Biochem. Biophys. 2015, 72, 275–282. [Google Scholar] [CrossRef]
- Yang, M.; Shan, X.; Zhou, X.; Qiu, T.; Zhu, W.; Ding, Y.; Shu, Y.; Liu, P. miR-1271 regulates cisplatin resistance of human gastric cancer cell lines by targeting IGF1R, IRS1, mTOR, and BCL2. Anticancer Agents Med. Chem. 2014, 14, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qu, X.; Li, C.; Fan, Y.; Che, X.; Wang, X.; Cai, Y.; Hu, X.; Liu, Y. miR-103/107 modulates multidrug resistance in human gastric carcinoma by downregulating Cav-1. Tumour Biol. 2015, 36, 2277–2785. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhu, M.; Zhou, X.; Huang, Z.; Zhu, J.; Xu, J.; Cheng, G.; Shu, Y.; Liu, P.; Zhu, W.; Wang, T. miR-20a enhances cisplatin resistance of human gastric cancer cell line by targeting NFKBIB. Tumour Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Zhang, Z.; Liu, Z.; Feng, B.; Ren, G.; Li, K.; Zhou, L.; Sun, Y.; Li, M.; Zhou, J.; et al. miR-508-5p regulates multidrug resistance of gastric cancer by targeting ABCB1 and ZNRD1. Oncogene 2013, 33, 3267–3276. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yang, Z.; Xia, L.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. Methylation of miR-129-5p CpG island modulates multi-drug resistance in gastric cancer by targeting ABC transporters. Oncotarget 2014, 5, 11552–11563. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chen, L.; Cao, X.; Ha, S.; Xie, X. Expression profiling and functional analysis of hsa-miR-125b and its target genes in drug-resistant cell line of human gastric cancer. Hereditas 2014, 36, 119–126. [Google Scholar] [PubMed]
- An, Y.; Zhang, Z.; Shang, Y.; Jiang, X.; Dong, J.; Yu, P.; Nie, Y.; Zhao, Q. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jin, W.; Jia, H.; Yan, J.; Zhang, G. miR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J. Exp. Clin. Cancer Res. 2015, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-L.; Shi, H.-J.; Wang, J.-P.; Tang, H.-S.; Cui, S.-Z. MiR-218 inhibits multidrug resistance (MDR) of gastric cancer cells by targeting Hedgehog/smoothened. Int. J. Clin. Exp. Pathol. 2015, 8, 6397–6406. [Google Scholar] [PubMed]
- Wu, H.; Huang, M.; Lu, M.; Zhu, W.; Shu, Y.; Cao, P.; Liu, P. Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer Chemother. Pharmacol. 2013, 71, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ru, J.; Zhang, J.; Zhu, L.; Liu, M.; Li, X.; Tang, H. miR-23a targets interferon regulatory factor 1 and modulates cellular proliferation and paclitaxel-induced apoptosis in gastric adenocarcinoma cells. PLoS ONE 2013, 8, e64707. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.; Strasser, A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.C.; Milner, A.E.; Gregory, C.D.; Jackman, A.L.; Aherne, G.W.; Hartley, J.A.; Dive, C.; Hickman, J.A. bcl-2 modulation of apoptosis induced by anticancer drugs: Resistance to thymidylate stress is independent of classical resistance pathways. Cancer Res. 1993, 53, 3321–3326. [Google Scholar] [PubMed]
- Han, Z.; Hong, L.; Han, Y.; Wu, K.; Han, S.; Shen, H.; Li, C.; Yao, L.; Qiao, T.; Fan, D. Phospho Akt mediates multidrug resistance of gastric cancer cells through regulation of P-gp, Bcl-2 and Bax. J. Exp. Clin. Cancer Res. 2007, 26, 261–268. [Google Scholar] [PubMed]
- Tapia, O.; Riquelme, I.; Leal, P.; Sandoval, A.; Aedo, S.; Weber, H.; Letelier, P.; Bellolio, E.; Villaseca, M.; Garcia, P.; et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014, 465, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-G.; Wang, Q.; Zhou, C.-Z.; Qiu, G.-Q.; Peng, Z.-H.; Tang, H.-M. Mutation analysis of tumor suppressor gene PTEN in patients with gastric carcinomas and its impact on PI3K/AKT pathway. Oncol. Rep. 2010, 24, 89–95. [Google Scholar] [PubMed]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Keniry, M.; Parsons, R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene 2008, 27, 5477–5485. [Google Scholar] [CrossRef] [PubMed]
- Denduluri, S.K.; Idowu, O.; Wang, Z.; Liao, Z.; Yan, Z.; Mohammed, M.K.; Ye, J.; Wei, Q.; Wang, J.; Zhao, L.; et al. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2015, 2, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Casa, A.J.; Dearth, R.K.; Litzenburger, B.C.; Lee, A.V.; Cui, X. The type I insulin-like growth factor receptor pathway: A key player in cancer therapeutic resistance. Front. Biosci. 2008, 13, 3273–3287. [Google Scholar] [CrossRef] [PubMed]
- Hodges, L.M.; Markova, S.M.; Chinn, L.W.; Gow, J.M.; Kroetz, D.L.; Klein, T.E.; Altman, R.B. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein). Pharmacogenet. Genom. 2011, 21, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Fromm, M.F. The influence of MDR1 polymorphisms on P-glycoprotein expression and function in humans. Adv. Drug Deliv. Rev. 2002, 54, 1295–1310. [Google Scholar] [CrossRef]
- Leschziner, G.D.; Andrew, T.; Pirmohamed, M.; Johnson, M.R. ABCB1 genotype and PGP expression, function and therapeutic drug response: A critical review and recommendations for future research. Pharmacogenomics J. 2007, 7, 154–179. [Google Scholar] [CrossRef] [PubMed]
- Marzolini, C.; Paus, E.; Buclin, T.; Kim, R.B. Polymorphisms in human MDR1 (P-glycoprotein): Recent advances and clinical relevance. Clin. Pharmacol. Ther. 2004, 75, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Modok, S.; Heyward, C.; Callaghan, R. P-glycoprotein retains function when reconstituted into a sphingolipid- and cholesterol-rich environment. J. Lipid Res. 2004, 45, 1910–1918. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Bae, E.; Lee, C.; Yoon, S.-S.; Chae, Y.S.; Ahn, K.-S.; Won, N.H. RNA interference-directed caveolin-1 knockdown sensitizes SN12CPM6 cells to doxorubicin-induced apoptosis and reduces lung metastasis. Tumour Biol. 2010, 31, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Meena, A.S.; Sharma, A.; Kumari, R.; Mohammad, N.; Singh, S.V.; Bhat, M.K. Inherent and acquired resistance to paclitaxel in hepatocellular carcinoma: molecular events involved. PLoS ONE 2013, 8, e61524. [Google Scholar]
- Zhu, L.-H.; Liu, T.; Tang, H.; Tian, R.-Q.; Su, C.; Liu, M.; Li, X. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010, 277, 3726–3734. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shao, X.; Meng, X.; Zhang, X.; Zhu, L.; Liu, S.; Lin, J.; Xiao, H. Genome-wide analysis of microRNA and mRNA expression signatures in hydroxycamptothecin-resistant gastric cancer cells. Acta Pharmacol. Sin. 2011, 32, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Kim, H.K.; Rettig, R.L.; Kim, J.; Lee, E.T.; Aprelikova, O.; Choi, I.J.; Munroe, D.J.; Green, J.E. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med. Genom. 2011, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wang, H.; Liu, R.; Li, H.; Ge, S.; Bai, M.; Deng, T.; Yao, G.; Ba, Y. miRNA27a is a biomarker for predicting chemosensitivity and prognosis in metastatic or recurrent gastric cancer. J. Cell. Biochem. 2014, 115, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, R.L.; Martins, I.; Davidson, B.L. Artificial microRNAs as siRNA shuttles: Improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009, 17, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Baumann, V.; Winkler, J. miRNA-based therapies: Strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014, 6, 1967–1984. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006, 441, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, J. Delivery systems for siRNA drug development in cancer therapy. Asian J. Pharm. Sci. 2015, 10. [Google Scholar] [CrossRef]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dai, Z.; Liang, Y.; Yin, M.; Ma, K.; He, M.; Ouyang, H.; Teng, C.-B. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Meng, X.; Meng, L.; Chang, N.; Xiong, J.; Cao, H.; Liang, Z. Small indels induced by CRISPR/Cas9 in the 5′ region of microRNA lead to its depletion and Drosha processing retardance. RNA Biol. 2014, 11, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.-T.; Zhou, N.; Huang, J.; Koirala, P.; Xu, M.; Fung, R.; Wu, F.; Mo, Y.-Y. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-T.; Chen, Y.-Q. Circulating miRNAs in cancer: From detection to therapy. J. Hematol. Oncol. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.-C.; Li, Q.-G.; Ding, X.-W.; Ding, Y.-T. Circulating microRNAs: Potential biomarkers for cancer. Int. J. Mol. Sci. 2011, 12, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Jiang, T.; Kang, X. Circulating microRNAs in cancer: Origin, function and application. J. Exp. Clin. Cancer Res. 2012, 31. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Expression | Target Genes 1 | Tested Drugs 2 | References |
|---|---|---|---|---|
| miR-200bc/429 | Down | BCL2, XIAP | VCR, CDDP, VP-16, ADR, 5-Fu | [32] |
| miR-181b | Down | BCL2 | VCR, CDDP, 5-Fu, VP-16, ADR, MMC | [33] |
| miR-497 | Down | BCL2, IGF1R, IRS1 | VCR, CDDP, VP-16, ADR, 5-Fu | [34,35] |
| miR-503 | Down | BCL2, IGF1R | VCR, CDDP | [36,37] |
| miR-143 | Down | BCL2, IGF1R, ERK5 * | CDDP, 5-Fu | [38,39] |
| miR-449a | Down | BCL2, CCDN1 | CDDP | [40] |
| miR-15b | Down | BCL2 | VCR, CDDP, VP-16, ADR, 5-Fu, MMC | [41] |
| miR-16 | Down | BCL2 | VCR, CDDP, VP-16, ADR, 5-Fu, MMC | [41] |
| miR-34a/b | Down | BCL2 NOTCH pathway *, HMGA2 | CDDP, DOX, DTX, GEM | [42] |
| miR-204 | Down | BCL2 | 5-Fu, L-OHP | [43] |
| miR-19a/b | Up | PTEN | CDDP, 5-Fu, ADR | [44] |
| miR-106a | Up | PTEN, RUNX3 | CDDP, ADR | [45,46] |
| miR-21 | Up | PTEN, ABCB1 * | CDDP, PTX, Trastuzumab | [47,48,49] |
| miR-1271 | Down | IGF1R, IRS1, MTOR BCL2 | CDDP | [50] |
| miR-103/107 | Down | CAV1 | DOX | [51] |
| miR-20a | Up | NFKBIB | CDDP | [52] |
| miR-145 | Down | IRS1 *, BACT * | 5-Fu | [39] |
| miR-508-5p | Down | ABCB1, ZNRD1 | VCR, CDDP, 5-Fu, ADR | [53] |
| miR-129-5p | Down | ABCB1 *, ABCC5 *, ABCG1 * | VCR, ADR, CDDP, 5-Fu | [54] |
| miR-125b | Down | MAPK pathway **, WNT pathway **, P53 pathway ** | 5-Fu | [55] |
| miR-23b-3p | Down | ATG12, HMGB2 | VCR, CDDP, 5-Fu | [56] |
| miR-223 | Up | FBXW7 | CDDP | [57] |
| miR-218 | Down | SMO, ABCB1 *, BCL2 * | ADR, 5- Fu, L-OHP | [58] |
| miR-34c-5p | Down | MAPT | PTX | [59] |
| miR-23a | Up | IRF1 | PTX | [60] |
| If These miRNAs Are UPREGULATED | Consider a Potential Treatment with |
| miR-200bc/429 | VCR, CDDP, VP-16, ADR |
| miR-181b | |
| miR-497 | |
| miR-15b | |
| miR-16 | |
| miR-508-5p | VCR, CDDP, 5-Fu, |
| miR-129-5p | |
| miR-23b-3p | |
| miR-503 | |
| If these miRNAs are DOWNREGULATED | Consider a potential treatment with |
| miR-19a/b | CDDP |
| miR-106a | |
| miR-21 | |
| miR-20a | |
| miR-223 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riquelme, I.; Letelier, P.; Riffo-Campos, A.L.; Brebi, P.; Roa, J.C. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int. J. Mol. Sci. 2016, 17, 424. https://doi.org/10.3390/ijms17030424
Riquelme I, Letelier P, Riffo-Campos AL, Brebi P, Roa JC. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. International Journal of Molecular Sciences. 2016; 17(3):424. https://doi.org/10.3390/ijms17030424
Chicago/Turabian StyleRiquelme, Ismael, Pablo Letelier, Angela L. Riffo-Campos, Priscilla Brebi, and Juan Carlos Roa. 2016. "Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer" International Journal of Molecular Sciences 17, no. 3: 424. https://doi.org/10.3390/ijms17030424
APA StyleRiquelme, I., Letelier, P., Riffo-Campos, A. L., Brebi, P., & Roa, J. C. (2016). Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. International Journal of Molecular Sciences, 17(3), 424. https://doi.org/10.3390/ijms17030424