UPLC-MS/MS-Based Profiling of Eicosanoids in RAW264.7 Cells Treated with Lipopolysaccharide
Abstract
:1. Introduction
2. Results
2.1. Effect of Lipopolysaccharide (LPS) on RAW264.7 Cells
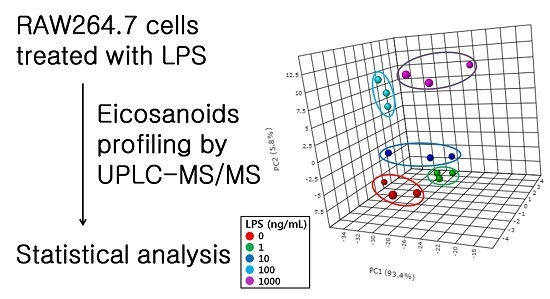
2.2. Profiling of Eicosanoids by Ultra Performance Liquid Chromatography Coupled to Tandem Mass Spectrometry (UPLC-MS/MS)
2.3. Quantification of Eicosanoids in LPS-Treated RAW264.7 Cells
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture and LPS Treatment
4.3. Nitric Oxide (NO) and Enzyme-Linked Immunosorbent Assays (ELISAs) Analyses
4.4. Sample Preparation
4.5. UPLC-MS/MS Conditions
4.6. Validation Study
4.7. Data Processing and Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LPS | Lipopolysaccharide |
| UPLC | Ultra performance liquid chromatography |
| MS | Mass spectrometry |
| PG | Prostaglandin |
| LT | Leukotriene |
| ELISA | Enzyme-linked immunosorbent assays |
| NO | Nitric oxide |
| TNF-α | Tumor necrosis factor-α |
| IL-6 | Interleukin-6 |
| MRM | Multiple reaction monitoring |
| CE | Collision energy |
| DP | Declustering potential |
| RSD | Relative standard deviation |
| LOD | The limit of detection |
| LOQ | The limit of quantification |
| IS | Internal standard |
| PCA | Principal component analysis |
| EDTA | Ethylenediaminetetraacetic acid |
References
- Sellmayer, A.; Koletzko, B. Long-chain polyunsaturated fatty acids and eicosanoids in infants—Physiological and pathophysiological aspects and open questions. Lipids 1999, 34, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.D. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science 2001, 294, 1871–1875. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Chiang, N.; van Dyke, T.E. Resolving inflammation: Dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 2008, 8, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Harizi, H.; Corcuff, J.B.; Gualde, N. Arachidonic-acid-derived eicosanoids: Roles in biology and immunopathology. Trends Mol. Med. 2008, 14, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.I.; Higgs, G.A. Eicosanoids and inflammation. J. Pathol. 1988, 156, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Libby, P. Inflammation and cardiovascular disease mechanisms. Am. J. Clin. Nutr. 2006, 83, 456S–460S. [Google Scholar] [PubMed]
- Hartge, M.M.; Unger, T.; Kintscher, U. The endothelium and vascular inflammation in diabetes. Diabetes Vasc. Dis. Res. 2007, 4, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Tasi, M.; Piliponsky, A.M. The development of allergic inflammation. Nature 2008, 454, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Lakoski, S.G.; Cushman, M.; Siscovick, D.S.; Blumenthal, R.S.; Palmas, W.; Burke, G.; Herrington, D.M. The relationship between inflammation, obesity and risk for hypertension in the Multi-Ethnic Study of Atherosclerosis (MESA). J. Hum. Hypertens. 2011, 25, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; DuBois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Ferreiro-Vera, C.; Mata-Granados, J.M.; Priego-Capote, F.; Quesada-Gomez, J.M.; Luque de Castro, M.D. Automated targeting analysis of eicosanoid inflammation biomarkers in human serum and in the exometabolome of stem cells by SPE-LC-MS/MS. Anal. Bioanal. Chem. 2011, 399, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Norwood, S.; Liao, J.; Hammock, B.D.; Yang, G.Y. Epoxyeicosatrienoic acids and soluble epoxide hydrolase: Potential therapeutic target for inflammation and its induced carcinogenesis. Am. J. Transl. Res. 2010, 2, 447–457. [Google Scholar] [PubMed]
- Birney, Y.; Redmond, E.M.; Sitzmann, J.V.; Cahill, P.A. Eicosanoids in cirrhosis and portal hypertension. Prostaglandins Other Lipid Mediat. 2003, 72, 3–18. [Google Scholar] [CrossRef]
- Harizi, H.; Grosset, C.; Gualde, N. Prostaglandin E2 Modulates dendritic cell function via EP2 and EP4 receptor subtypes. J. Leukoc. Biol. 2003, 73, 1–8. [Google Scholar] [CrossRef]
- Miller, D.K.; Sadowski, S.; DeSousa, D.; Maycock, A.L.; Lombardo, D.L.; Young, R.N.; Hayes, E.C. Development of enzyme-linked immunosorbent assays for measurement of leukotrienes and prostaglandins. J. Immunol. Methods 1985, 81, 169–185. [Google Scholar] [CrossRef]
- Mazid, M.A.; Nishimura, K.; Nagao, K.; Jisaka, M.; Nagaya, T.; Yokota, K. Development of enzyme-linked immunosorbent assay for prostaglandin D2 using the stable isosteric analogue as a hapten mimic and its application. Prostaglandins Other Lipid Mediat. 2007, 83, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Alber, D.; Moussard, C.; Toubin, M.; Henry, J.C.; Ottignon, Y.; Deschamps, J.P. Gas chromatographic/mass spectrometric quantitative analysis of eicosanoids in human oesophageal mucosa. Biomed. Environ. Mass Spectrom. 1988, 16, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Deems, R.; Buczynski, M.W.; Bowers-Gentry, R.; Harkewicz, R.; Dennis, E.A. Detection and quantitation of eicosanoids via high performance liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol. 2007, 432, 59–82. [Google Scholar] [PubMed]
- Wan, G.H.; Yan, D.C.; Tseng, H.Y.; Lee, J.T.; Lin, Y.W. Using high-performance liquid chromatography with UV detector to quantify exhaled leukotriene B4 level in nonatopic adults. J. Formos. Med. Assoc. 2014, 113, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Kita, Y.; Takahashi, T.; Uozumi, N.; Shimizu, T. A multiplex quantitation method for eicosanoids and platelet-activating factor using column-switching reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Biochem. 2005, 342, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Martin-Venegas, R.; Jauregui, O.; Moreno, J.J. Liquid chromatography-tandem mass spectrometry analysis of eicosanoids and related compounds in cell models. J. Chromatogr. B 2014, 964, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Paolo, M. LC/MS/MS analysis of leukotriene B4 and other eicosanoids in exhaled breath condensate for assessing lung inflammation. J. Chromatogr. B 2009, 877, 1272–1280. [Google Scholar]
- Kingsley, P.J.; Marnett, L.J. LC-MS-MS analysis of neutral eicosanoids. Methods Enzymol. 2007, 433, 91–112. [Google Scholar] [PubMed]
- Wang, Y.; Armando, A.M.; Quehenberger, O.; Yan, C.; Dennis, E.A. Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J. Chromatogr. A 2014, 1359, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Kortz, L.; Dorow, J.; Ceglarek, U. Liquid chromatography-tandem mass spectrometry for the analysis of eicosanoids and related lipids in human biological matrices: A review. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 964, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Norris, P.C.; Dennis, E.A. A lipid perspective on inflammatory macrophage eicosanoid signaling. Adv. Biol. Regul. 2014, 54, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Trebino, C.E.; Eskra, J.D.; Wachtmann, T.S.; Perez, J.R.; Carty, T.J.; Audoly, L.P. Redirection of eicosanoid metabolism in mPGES-1-deficient macrophages. J. Biol. Chem. 2005, 280, 16579–16585. [Google Scholar] [CrossRef] [PubMed]
- Bromfield, J.J.; Sheldon, I.M. Lipopolysaccharide initiates inflammation in bovine granulosa cells via the TLR4 pathway and perturbs oocyte meiotic progression in vitro. Endocrinology 2011, 152, 5029–5040. [Google Scholar] [CrossRef] [PubMed]
- Djoko, B.; Chiou, R.Y.Y.; Shee, J.J.; Liu, Y.W. Characterization of immunological activities of peanut stilbenoids, arachidin-1, piceatannol, and resveratrol on lipopolysaccharide-induced inflammation of RAW264.7 macrophages. J. Agric. Food Chem. 2007, 55, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.K.; Vallyathan, V.; Lewis, D.M. Evidence for lipopolysaccharide-induced differentiation of RAW264.7 murine macrophage cell line into dendritic like cells. J. Biosci. 2003, 28, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.R.; Han, D.Y.; Park, K.I.; Park, H.S.; Cho, Y.B.; Lee, H.J.; Lee, W.S.; Ryu, C.H.; Ha, Y.L.; Lee, D.H.; et al. Suppressive effect on lipopolysaccharide-induced proinflammatory mediators by Citrus aurantium L. in macrophage RAW264.7 cells via NF-κB signal pathway. Evid. Based Complement. Altern. Med. 2011, 2011, 1–12. [Google Scholar]
- Dumlao, D.S.; Buczynski, M.W.; Norris, P.C.; Harkewicz, R.; Dennis, E.A. High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines. Biochim. Biophys. Acta 2011, 1811, 724–736. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Halouska, S.; Powers, R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal. Biochem. 2013, 433, 102–104. [Google Scholar] [CrossRef] [PubMed]
- Nyamundanda, G.; Brennan, L.; Gormley, I.C. Probabilistic principal component analysis for metabolomic data. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Almer, G.; Teismann, P.; Stevic, Z.; Halaschek-Wiener, J.; Deecke, L.; Kostic, V.; Przedborski, S. Increased levels of the pro-inflammatory prostaglandin PGE2 in CSF from ALS patients. Neurology 2001, 58, 1277–1279. [Google Scholar] [CrossRef]
- Kuehl, F.A.; Egan, R.W. Prostaglandins, arachidonic acid, and inflammation. Science 1980, 210, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Higashi, N.; Mita, H.; Ono, E.; Fukutomi, Y.; Yamaguchi, H.; Kajiwara, K.; Tanimoto, H.; Sekiya, K.; Akiyama, K.; Taniguchi, M. Profile of eicosanoid generation in aspirin-intolerant asthma and anaphylaxis assessed by new biomarkers. J. Allergy Clin. Immunol. 2010, 125, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Ecker, J. Profiling eicosanoids and phospholipids using LC-MS/MS: Principles and recent applications. J. Sep. Sci. 2012, 35, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Drozdovszky, O.; Barta, I.; Antus, B. Sputum eicosanoid profiling in exacerbations of chronic obstructive pulmonary disease. Respiration 2014, 87, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Mal, M.; Koh, P.K.; Cheah, P.Y.; Chan, E.C.Y. Ultra-pressure liquid chromatography/tandem mass spectrometry targeted profiling of arachidonic acid and eicosanoids in human colorectal cancer. Rapid Commun. Mass Spectrom. 2011, 25, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Rago, B.; Fu, C. Development of a high-throughput ultra performance liquid chromatography-mass spectrometry assay to profile 18 eicosanoids as exploratory biomarkers for atherosclerotic diseases. J. Chromatogr. B 2013, 936, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, K.; Gross, R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012, 31, 134–178. [Google Scholar] [CrossRef] [PubMed]
- MetaboAnalyst 3.0—A Comprehensive Tool Suite for Meabolomic Data Analysis. Avaliable online: http://www.metaboanalyst.ca (accessed on 1 April 2016).







| Compounds | Abbreviation | Ion Mode | MRM Transitions | DP | CE | |
|---|---|---|---|---|---|---|
| Q1 | Q3 | |||||
| Prostaglandin E2-d4 | PGE2-d4 | Negative | 355 | 275 | −50 | −25 |
| Prostaglandin D2-d4 | PGD2-d4 | Negative | 355 | 275 | −50 | −25 |
| Leukotriene B4-d4 | LTB4-d4 | Negative | 339 | 197 | −70 | −22 |
| 14,15 Epoxy-eicosatrienoic acid-d11 | 14,15 EET-d11 | Negative | 330 | 202 | −50 | −15 |
| 5-Hydroxy-eicosatetraenoic acid-d8 | 5(S)-HETE-d8 | Negative | 327 | 116 | −50 | −20 |
| Arachidonic acid-d8 | AA-d8 | Negative | 311 | 267 | −80 | −20 |
| Eicosanoids | RT (min) | RSD (n = 6) (%) | Correlation (R2) | Linear Range (pg) | LOD (pg) | LOQ (pg) | |
|---|---|---|---|---|---|---|---|
| RT | Peak Area | ||||||
| PGE2-d4 | 8.06 | 0.25 | 2.85 | 0.9978 | 30–10,000 | 3 | 30 |
| PGD2-d4 | 8.43 | 0 | 4.44 | 0.9982 | 30–10,000 | 3 | 30 |
| LTB4-d4 | 12.77 | 0.20 | 5.39 | 0.9969 | 100–10,000 | 60 | 100 |
| 14,15 EET-d11 | 17.70 | 0 | 6.08 | 0.9964 | 10,000–100,000 | 3000 | 10,000 |
| 5(S)-HETE-d8 | 17.85 | 0 | 5.01 | 0.9972 | 30–10,000 | 6 | 30 |
| AA-d8 | 21.53 | 0 | 7.84 | 0.9959 | 3–6000 | 0.1 | 3 |
| No. | Compound Name | Abbreviation | MRM Transitions | DP | CE | RT (min) | IS | Alteration by LPS Treatment | |
|---|---|---|---|---|---|---|---|---|---|
| Q1 | Q3 | ||||||||
| 1 | 12S-hydroxy-heptadecatrienoic acid | 12-HHT | 279 | 163 | −30 | −30 | 14.3 | 5(S)HETE-d8 | Up |
| 2 | 13-hydroxy-g-octadecatrienoic acid | 13-HOTre-g | 293 | 193 | −70 | −20 | 15.5 | 5(S)HETE-d8 | Up |
| 3 | 9-hydroxy-octadecadienoic acid | 9-HODE | 295 | 171 | −60 | −25 | 16.6 | 5(S)HETE-d8 | Up |
| 4 | 13-hydroxy-octadecadienoic acid | 13-HODE | 295 | 195 | −60 | −25 | 16.3 | 5(S)HETE-d8 | – |
| 5 | 9,10-hydroxy-octadecadienoic acid | 9,10-diHOME | 313 | 201 | −60 | −30 | 14.2 | 5(S)HETE-d8 | – |
| 6 | 12,13-hydroxy-octadecadienoic acid | 12,13-diHOME | 313 | 183 | −60 | −30 | 13.5 | 5(S)HETE-d8 | – |
| 7 | 9-hydroxy-eicosapentaenoic acid | 9-HEPE | 317 | 149 | −75 | −20 | 15.7 | 5(S)HETE-d8 | Up |
| 8 | 5-hydroxy-eicosapentaenoic acid | 5-HEPE | 317 | 115 | −40 | −17 | 16.6 | 5(S)HETE-d8 | Down |
| 9 | 15-hydroxy-eicosapentaenoic acid | 15-HEPE | 317 | 219 | −60 | −20 | 15.5 | 5(S)HETE-d8 | Up |
| 10 | 8-hydroxy-eicosapentaenoic acid | 8-HEPE | 317 | 127 | −70 | −25 | 15.5 | 5(S)HETE-d8 | – |
| 11 | 11-hydroxy-eicosapentaenoic acid | 11-HEPE | 317 | 121 | −70 | −24 | 15.7 | 5(S)HETE-d8 | Up |
| 12 | 12-hydroxy-eicosapentaenoic acid | 12-HEPE | 317 | 179 | −70 | −20 | 15.9 | 5(S)HETE-d8 | – |
| 13 | 18-hydroxy-eicosapentaenoic acid | 18-HEPE | 317 | 215 | −60 | −20 | 15.1 | 5(S)HETE-d8 | – |
| 14 | 11-hydroxy-eicosatetraenoic acid | 11-HETE | 319 | 167 | −60 | −20 | 17.1 | 5(S)HETE-d8 | Up |
| 15 | 9-hydroxy-eicosatetraenoic acid | 9-HETE | 319 | 151 | −60 | −20 | 17.0 | 5(S)HETE-d8 | Up |
| 16 | 5-hydroxy-eicosatetraenoic acid | 5-HETE | 319 | 115 | −60 | −20 | 17.9 | 5(S)HETE-d8 | Down |
| 17 | 8-hydroxy-eicosatetraenoic acid | 8-HETE | 319 | 155 | −60 | −20 | 17.3 | 5(S)HETE-d8 | – |
| 18 | 15-hydroxy-eicosatetraenoic acid | 15-HETE | 319 | 219 | −50 | −15 | 16.7 | 5(S)HETE-d8 | Up |
| 19 | 12-hydroxy-eicosatetraenoic acid | 12-HETE | 319 | 179 | −60 | −20 | 17.3 | 5(S)HETE-d8 | Up |
| 20 | 18-hydroxy-eicosatetraenoic acid | 18-HETE | 319 | 261 | −80 | −25 | 15.9 | 5(S)HETE-d8 | – |
| 21 | 17-hydroxy-eicosatetraenoic acid | 17-HETE | 319 | 247 | −80 | −25 | 15.9 | 5(S)HETE-d8 | – |
| 22 | 16-hydroxy-eicosatetraenoic acid | 16-HETE | 319 | 189 | −80 | −25 | 16.0 | 5(S)HETE-d8 | – |
| 23 | 5-hydroxy-eicosatrienoic acid | 5-HETrE | 321 | 115 | −70 | −19 | 19.1 | 5(S)HETE-d8 | Down |
| 24 | 15-hydroxy-eicosatrienoic acid | 15-HETrE | 321 | 221 | −70 | −21 | 17.4 | 5(S)HETE-d8 | Up |
| 25 | 5,6-dihydroxy-eicosatrienoic acid | 5,6-DHET | 337 | 145 | −75 | −25 | 16.5 | 5(S)HETE-d8 | Down |
| 26 | 8,9-dihydroxy-eicosatrienoic acid | 8,9-DHET | 337 | 127 | −60 | −30 | 15.5 | 5(S)HETE-d8 | – |
| 27 | 11,12-dihydroxy-eicosatrienoic acid | 11,12-DHET | 337 | 167 | −60 | −25 | 15.8 | 5(S)HETE-d8 | Up |
| 28 | 8-hydroxy-docosahexaenoic acid | 8-HDoHE | 343 | 109 | −70 | −20 | 17.5 | 5(S)HETE-d8 | – |
| 29 | 7-hydroxy-docosahexaenoic acid | 7-HDoHE | 343 | 141 | −60 | −18 | 17.3 | 5(S)HETE-d8 | – |
| 30 | 4-hydroxy-docosahexaenoic acid | 4-HDoHE | 343 | 101 | −70 | −17 | 18.2 | 5(S)HETE-d8 | – |
| 31 | 10-hydroxy-docosahexaenoic acid | 10-HDoHE | 343 | 181 | −60 | −17 | 16.9 | 5(S)HETE-d8 | – |
| 32 | 11-hydroxy-docosahexaenoic acid | 11-HDoHE | 343 | 149 | −60 | −19 | 17.0 | 5(S)HETE-d8 | Up |
| 33 | 13-hydroxy-docosahexaenoic acid | 13-HDoHE | 343 | 221 | −60 | −17 | 16.7 | 5(S)HETE-d8 | Up |
| 34 | 16-hydroxy-docosahexaenoic acid | 16-HDoHE | 343 | 233 | −75 | −19 | 16.5 | 5(S)HETE-d8 | Up |
| 35 | 20-hydroxy-docosahexaenoic acid | 20-HDoHE | 343 | 241 | −60 | −20 | 16.3 | 5(S)HETE-d8 | – |
| 36 | 17-hydroxy-docosahexaenoic acid | 17-HDoHE | 343 | 245 | −60 | −20 | 16.5 | 5(S)HETE-d8 | Up |
| 37 | 14-hydroxy-docosahexaenoic acid | 14-HDoHE | 343 | 205 | −60 | −18 | 16.7 | 5(S)HETE-d8 | – |
| 38 | Arachidonic acid | AA | 303 | 259 | −80 | −20 | 21.6 | AA-d8 | Down |
| 39 | Eicosapentaenoic acid | EPA | 301 | 257 | −65 | −15 | 20.4 | AA-d8 | – |
| 40 | Adrenic acid | ADA | 331 | 287 | −80 | −20 | 22.3 | AA-d8 | – |
| 41 | Dohexacosaenoic acid | DHA | 327 | 283 | −95 | −20 | 21.3 | AA-d8 | – |
| 42 | 13-oxo-octadecadienoic acid | 13-oxoODE | 293 | 113 | −70 | −30 | 16.6 | 14,15 EET-d11 | – |
| 43 | 9,10-epoxy-octadecenoic acid | 9,10-EpOME | 295 | 171 | −60 | −25 | 18.4 | 14,15 EET-d11 | – |
| 44 | 12,13-epoxy-octadecenoic acid | 12,13-EpOME | 295 | 195 | −60 | −25 | 18.1 | 14,15 EET-d11 | – |
| 45 | 5-5-oxo-eicosatetraenoic acid | 5-oxoETE | 317 | 203 | −40 | −25 | 18.3 | 14,15 EET-d11 | – |
| 46 | 15-5-oxo-eicosatetraenoic acid | 15-oxoETE | 317 | 113 | −40 | −25 | 16.7 | 14,15 EET-d11 | – |
| 47 | 11,12-epoxy-eicosatrienoic acid | 11,12-EET | 319 | 167 | −60 | −20 | 18.6 | 14,15 EET-d11 | Down |
| 48 | 14,15-epoxy-eicosatrienoic acid | 14,15-EET | 319 | 219 | −50 | −15 | 18.1 | 14,15 EET-d11 | – |
| 49 | 5,6-epoxy-eicosatrienoic acid | 5,6-EET | 319 | 191 | −30 | −20 | 18.8 | 14,15 EET-d11 | Down |
| 50 | 15-oxo-eicosadienoic acid | 15-oxoEDE | 321 | 113 | −100 | −32 | 18.1 | 14,15 EET-d11 | Up |
| 51 | Hepoxilin B3 | HXB3 | 335 | 183 | −40 | −20 | 15.5 | 14,15 EET-d11 | – |
| 52 | 19,20-epoxy Docosapentaenoic acid | 19,20-EpDPE | 343 | 241 | −60 | −20 | 17.6 | 14,15 EET-d11 | – |
| 53 | Lipoxin B4 | LXB4 | 351 | 221 | −80 | −25 | 8.4 | LTB4-d4 | Up |
| 54 | 20-carboxy-Leukotriene B4 | 20cooh LTB4 | 365 | 195 | −60 | −25 | 6.4 | LTB4-d4 | – |
| 55 | 15-deoxy-Prostaglandin A2 or 15-deoxy-Δ12,14-PGJ2 | 15d-PGA2 or 15d-PGJ2 | 315 | 271 | −50 | −15 | 15.2 | PGE2-d4 | Up |
| 56 | Tetranor-Prostanglin F Metabolite | tetranor-PGFM | 329 | 293 | −40 | −25 | 3.1 | PGE2-d4 | – |
| 57 | Prostaglandin A2 or Prostaglandin B2 or Prostaglandin J2 | PGA2 or PGB2 or PGJ2 | 333 | 271 | −30 | −20 | 10.5 | PGE2-d4 | Up |
| 58 | 15-deoxy-Δ12,14-PGD2 | 15d-PGD2 | 333 | 271 | −30 | −20 | 12.8 | PGE2-d4 | Up |
| 59 | Prostaglandin D3 | PGD3 | 349 | 269 | −55 | −25 | 7.4 | PGE2-d4 | Up |
| 60 | Prostaglandin E3 | PGE3 | 349 | 269 | −55 | −25 | 7.1 | PGE2-d4 | Up |
| 61 | 15-keto-Prostaglandin E2 | 15k PGE2 | 349 | 113 | −35 | −30 | 8.3 | PGE2-d4 | – |
| 62 | Prostaglandin K2 | PGK2 | 349 | 205 | −50 | −30 | 8.3 | PGE2-d4 | Up |
| 63 | 15-keto-Prostaglandin F2 | 15k PGF2 | 351 | 113 | −40 | −35 | 8.6 | PGE2-d4 | Up |
| 64 | Prostaglandin E2 | PGE2 | 351 | 271 | −50 | -25 | 8.1 | PGE2-d4 | Up |
| 65 | Prostaglandin D2 | PGD2 | 351 | 271 | −50 | −25 | 8.4 | PGE2-d4 | Up |
| 66 | 13,14-dihydro-15-keto Prostaglandin E2 | dhk PGE2 | 351 | 207 | −40 | −25 | 8.4 | PGE2-d4 | Up |
| 67 | 13,14-dihydro-15-keto Prostaglandin D2 | dhk PGD2 | 351 | 207 | −40 | −25 | 9.3 | PGE2-d4 | Up |
| 68 | Prostaglandin F2α | PGF2α | 353 | 193 | −50 | −30 | 8.5 | PGE2-d4 | Up |
| 69 | 15-keto-Prostaglandin F1α | 15k PGF1α | 353 | 113 | −50 | −35 | 3.1 | PGE2-d4 | – |
| 70 | 11β-13,14-dihydro-15-keto-Prostaglandin F2α | 11β dhk PGF2α | 353 | 113 | −50 | −35 | 9.3 | PGE2-d4 | Up |
| 71 | Prostaglandin E1 | PGE1 | 353 | 273 | −55 | −25 | 8.1 | PGE2-d4 | Up |
| 72 | Prostaglandin D1 | PGD1 | 353 | 273 | −55 | −25 | 8.5 | PGE2-d4 | Up |
| 73 | Prostaglandin F1α | PGF1α | 355 | 293 | −75 | −30 | 8.2 | PGE2-d4 | Up |
| 74 | 13,14-dihydro-Prostaglandin F2α | dh PGF2α | 355 | 275 | −40 | −25 | 8.9 | PGE2-d4 | – |
| 75 | Dihomo-Prostaglandin J2 | Dihomo-PGJ2 | 361 | 299 | −55 | −25 | 13.0 | PGE2-d4 | Up |
| 76 | Dihomo-15-deoxy-Prostaglandin J2 | Dihomo-15d PGJ2 | 361 | 299 | −55 | −25 | 14.4 | PGE2-d4 | Up |
| 77 | Thromboxane B3 | TXB3 | 367 | 169 | −50 | −25 | 6.4 | PGE2-d4 | – |
| 78 | Dihomo-Prostaglandin F2α | Dihomo-PGF2α | 381 | 221 | −75 | −35 | 10.0 | PGE2-d4 | Up |
| 79 | Dihomo-Prostaglandin D2 | Dihomo-PGD2 | 379 | 299 | −65 | −30 | 10.4 | PGE2-d4 | Up |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.W.; Mok, H.J.; Lee, D.-Y.; Park, S.C.; Ban, M.S.; Choi, J.; Park, C.G.; Ahn, Y.-S.; Kim, K.P.; Kim, H.D. UPLC-MS/MS-Based Profiling of Eicosanoids in RAW264.7 Cells Treated with Lipopolysaccharide. Int. J. Mol. Sci. 2016, 17, 508. https://doi.org/10.3390/ijms17040508
Lee JW, Mok HJ, Lee D-Y, Park SC, Ban MS, Choi J, Park CG, Ahn Y-S, Kim KP, Kim HD. UPLC-MS/MS-Based Profiling of Eicosanoids in RAW264.7 Cells Treated with Lipopolysaccharide. International Journal of Molecular Sciences. 2016; 17(4):508. https://doi.org/10.3390/ijms17040508
Chicago/Turabian StyleLee, Jae Won, Hyuck Jun Mok, Dae-Young Lee, Seung Cheol Park, Myeong Soon Ban, Jehun Choi, Chun Geon Park, Young-Sup Ahn, Kwang Pyo Kim, and Hyung Don Kim. 2016. "UPLC-MS/MS-Based Profiling of Eicosanoids in RAW264.7 Cells Treated with Lipopolysaccharide" International Journal of Molecular Sciences 17, no. 4: 508. https://doi.org/10.3390/ijms17040508
APA StyleLee, J. W., Mok, H. J., Lee, D.-Y., Park, S. C., Ban, M. S., Choi, J., Park, C. G., Ahn, Y.-S., Kim, K. P., & Kim, H. D. (2016). UPLC-MS/MS-Based Profiling of Eicosanoids in RAW264.7 Cells Treated with Lipopolysaccharide. International Journal of Molecular Sciences, 17(4), 508. https://doi.org/10.3390/ijms17040508