Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures
Abstract
:1. Introduction
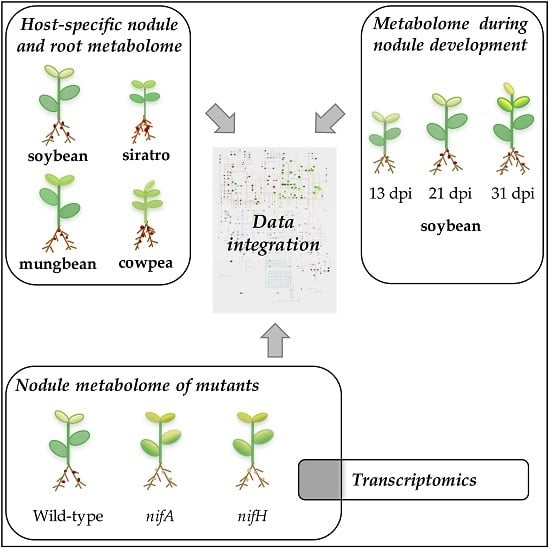
2. Results and Discussion
2.1. Metabolomic Analysis of B. diazoefficiens Root Nodules Identifies a Core Nodule Metabolome
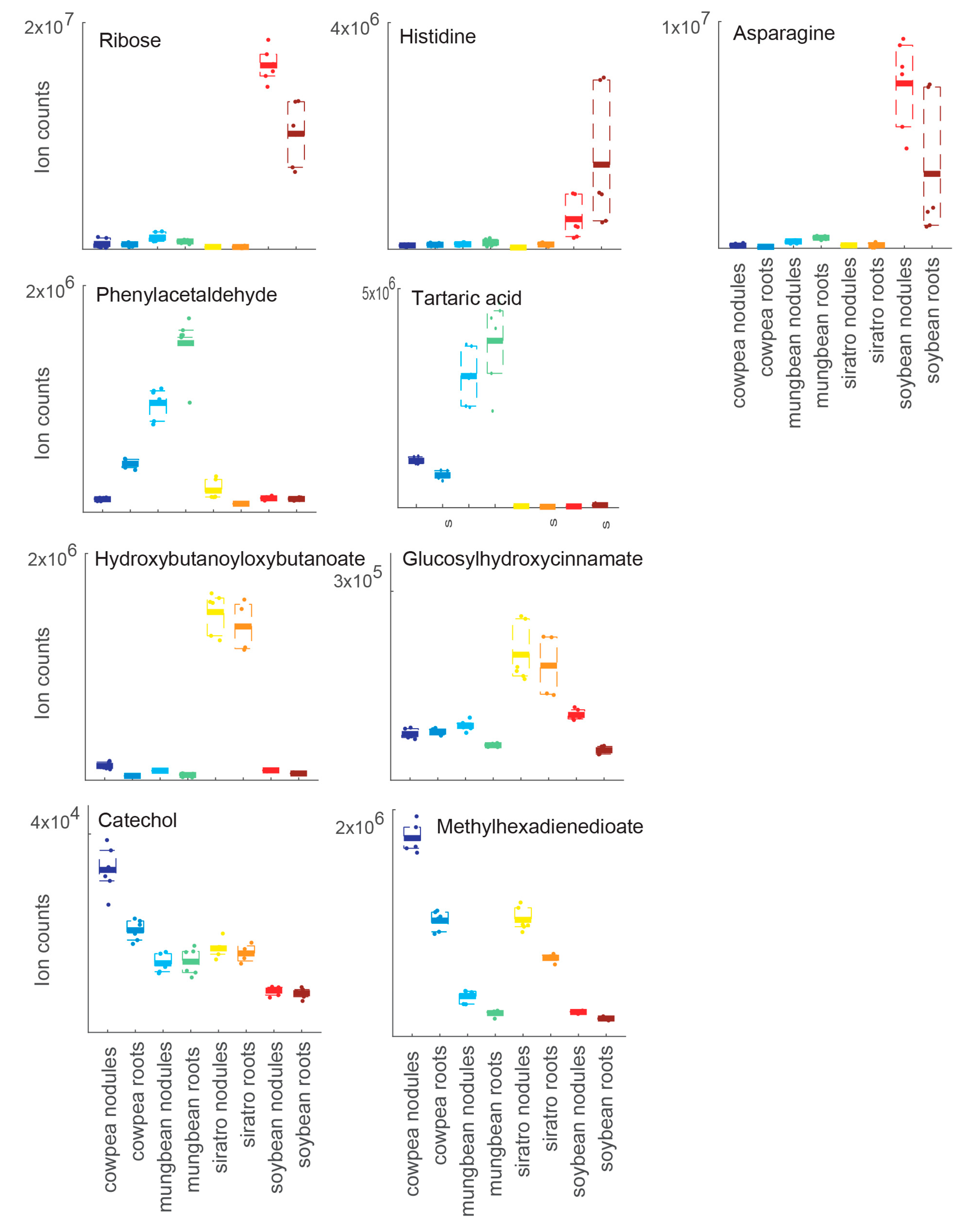
2.2. Host-Specific Nodule and Root Metabolome
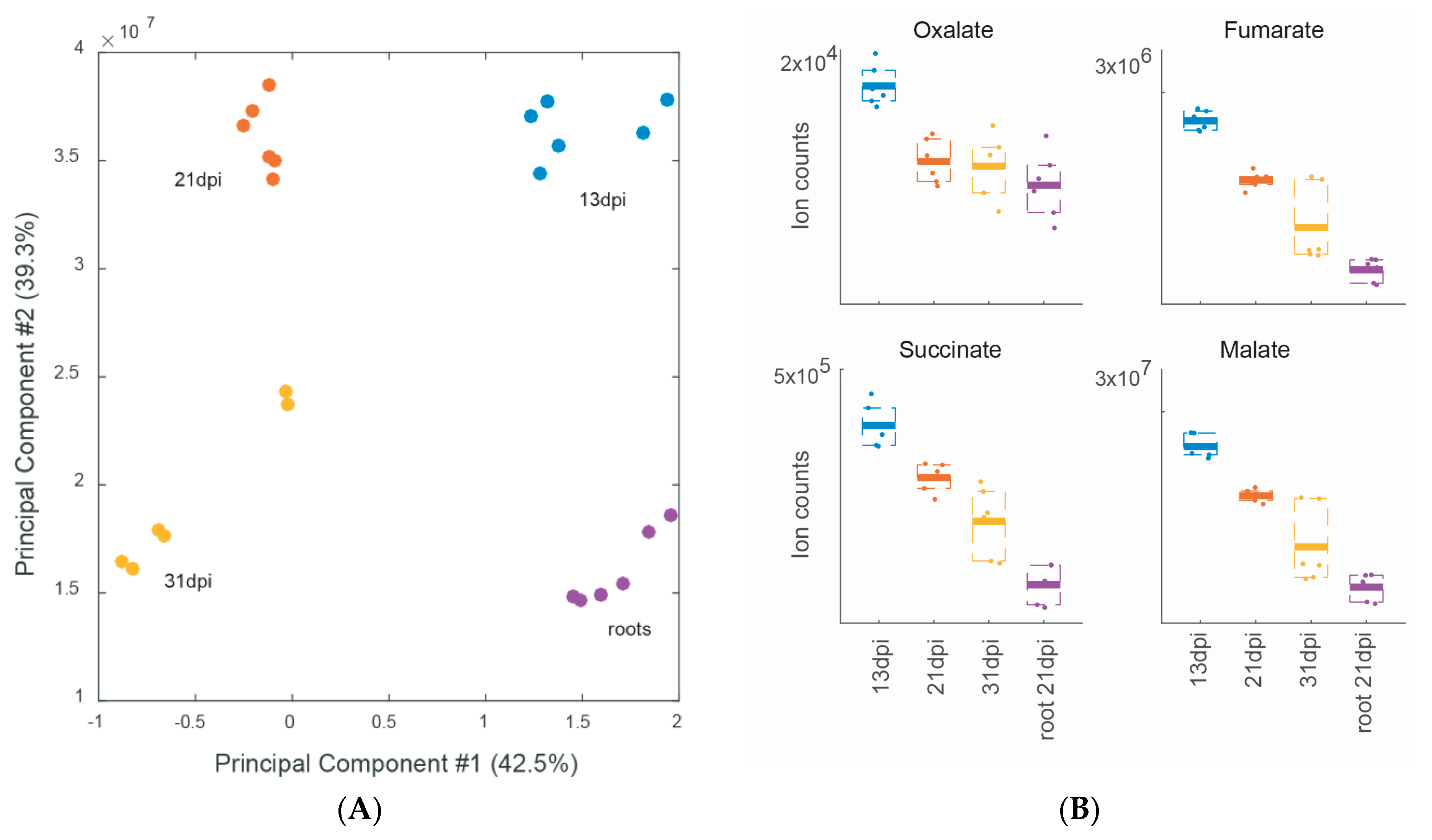
2.3. Metabolite Profiling during Different Stages of Soybean Nodule Development
2.4. Metabolomic Analysis of a B. diazoefficiens nifA and nifH Mutant
3. Materials and Methods
3.1. Bradyrhizobium Diazoefficiens Strains and Plant Growth
3.2. Plant Harvesting and Metabolite Extraction
3.3. Data Analysis
3.4. Transcriptome Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Göttfert, M.; Grob, P.; Hennecke, H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 1990, 87, 2680–2684. [Google Scholar] [CrossRef] [PubMed]
- Long, S.R. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 2001, 125, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Downie, J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Masson-Boivin, C.; Giraud, E.; Perret, X.; Batut, J. Establishing nitrogen-fixing symbiosis with legumes: How many Rhizobium recipes? Trends Microbiol. 2009, 17, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Prell, J.; White, J.P.; Bourdes, A.; Bunnewell, S.; Bongaerts, R.J.; Poole, P.S. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl. Acad. Sci. USA 2009, 106, 12477–12482. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.J.; Hood, G.A.; Poole, P.S. What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 2012, 60, 325–389. [Google Scholar] [PubMed]
- Udvardi, M.K.; Day, D.A. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 493–523. [Google Scholar] [CrossRef] [PubMed]
- Clarke, V.C.; Loughlin, P.C.; Day, D.A.; Smith, P.M. Transport processes of the legume symbiosome membrane. Front. Plant Sci. 2014, 5, 699. [Google Scholar] [CrossRef] [PubMed]
- Prell, J.; Poole, P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006, 14, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.; Poole, P.S. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.F. Key roles of microsymbiont amino acid metabolism in rhizobia-legume interactions. Crit. Rev. Microbiol. 2015, 41, 411–451. [Google Scholar] [CrossRef] [PubMed]
- Sciotti, M.A.; Chanfon, A.; Hennecke, H.; Fischer, H.M. Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic genes in Bradyrhizobium japonicum. J. Bacteriol. 2003, 185, 5639–5642. [Google Scholar] [CrossRef] [PubMed]
- Poole, P.; Allaway, D. Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 2000, 43, 117–163. [Google Scholar] [PubMed]
- Lodwig, E.; Poole, P. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 2003, 22, 37–78. [Google Scholar] [CrossRef]
- Gage, D.J. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 2004, 68, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Baral, B.; Teixeira da Silva, J.A.; Izaguirre-Mayoral, M.L. Early signaling, synthesis, transport and metabolism of ureides. J. Plant Physiol. 2016, 193, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.M.; Hennecke, H. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, N.; Ahrens, C.H.; Knief, C.; Qeli, E.; Koch, M.; Fischer, H.M.; Vorholt, J.A.; Hennecke, H.; Pessi, G. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 2010, 10, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Delmotte, N.; Rehrauer, H.; Vorholt, J.A.; Pessi, G.; Hennecke, H. Rhizobial adaptation to hosts, a new facet in the legume root-nodule symbiosis. Mol. Plant Microbe Interact. 2010, 23, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Pessi, G.; Friberg, M.; Weber, C.; Rusca, N.; Lindemann, A.; Fischer, H.M.; Hennecke, H. Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genom. 2007, 278, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, A.; Moser, A.; Pessi, G.; Hauser, F.; Friberg, M.; Hennecke, H.; Fischer, H.M. New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J. Bacteriol. 2007, 189, 8928–8943. [Google Scholar] [CrossRef] [PubMed]
- Mesa, S.; Hauser, F.; Friberg, M.; Malaguti, E.; Fischer, H.M.; Hennecke, H. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J. Bacteriol. 2008, 190, 6568–6579. [Google Scholar] [CrossRef] [PubMed]
- Brechenmacher, L.; Lei, Z.; Libault, M.; Findley, S.; Sugawara, M.; Sadowsky, M.J.; Sumner, L.W.; Stacey, G. Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol. 2010, 153, 1808–1822. [Google Scholar] [CrossRef] [PubMed]
- Vauclare, P.; Bligny, R.; Gout, E.; Widmer, F. An overview of the metabolic differences between Bradyrhizobium japonicum 110 bacteria and differentiated bacteroids from soybean (Glycine max) root nodules: An in vitro 13C-and 31P-nuclear magnetic resonance spectroscopy study. FEMS Microbiol. Lett. 2013, 343, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Fuhrer, T.; Heer, D.; Begemann, B.; Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection–Time-of-flight mass spectrometry. Anal. Chem. 2011, 83, 7074–7080. [Google Scholar] [CrossRef] [PubMed]
- Fuhrer, T.; Zamboni, N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015, 31, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Taté, R.; Riccio, A.; Caputo, E.; Iaccarino, M.; Patriarca, E.J. The Rhizobium etli metZ gene is essential for methionine biosynthesis and nodulation of Phaseolus vulgaris. Mol. Plant Microbe Interact. 1999, 12, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Desbrosses, G.G.; Kopka, J.; Udvardi, M.K. Lotus japonicus metabolic profiling. Development of gas chromatography–mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol. 2005, 137, 1302–1318. [Google Scholar] [CrossRef] [PubMed]
- Barsch, A.; Tellstrom, V.; Patschkowski, T.; Kuster, H.; Niehaus, K. Metabolite profiles of nodulated alfalfa plants indicate that distinct stages of nodule organogenesis are accompanied by global physiological adaptations. Mol. Plant Microbe Interact. 2006, 19, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Colebatch, G.; Desbrosses, G.; Ott, T.; Krusell, L.; Montanari, O.; Kloska, S.; Kopka, J.; Udvardi, M.K. Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J. 2004, 39, 487–512. [Google Scholar] [CrossRef] [PubMed]
- Salminen, S.O.; Streeter, J.G. Involvement of glutamate in the respiratory metabolism of Bradyrhizobium japonicum bacteroids. J. Bacteriol. 1987, 169, 495–499. [Google Scholar] [PubMed]
- Shelp, B.J.; Dasilva, M.C. Distribution and metabolism of xylem-borne ureido and amino-compounds in developing soybean shoots. Plant Physiol. 1990, 94, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, N.; Nishiwaki, T.; Mizukoshi, K.; Minagawa, R.; Takahashi, Y.; Chinushi, T.; Ohyama, T. Amino-acid-composition in xylem sap of soybean related to the evaluation of N2 fixation by the relative ureide method. Soil Sci. Plant Nutr. 1995, 41, 95–102. [Google Scholar] [CrossRef]
- Todd, C.D.; Tipton, P.A.; Blevins, D.G.; Piedras, P.; Pineda, M.; Polacco, J.C. Update on ureide degradation in legumes. J. Exp. Bot. 2006, 57, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Atkins, C.A. Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol. 2002, 128, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Katahira, R.; Ashihara, H. Metabolism of purine nucleosides and bases in suspension-cultured Arabidopsis thaliana cells. Eur. Chem. Bull. 2014, 3, 925–934. [Google Scholar]
- Koch, M.; Delmotte, N.; Ahrens, C.H.; Omasits, U.; Schneider, K.; Danza, F.; Padhi, B.; Murset, V.; Braissant, O.; Vorholt, J.A.; et al. A link between arabinose utilization and oxalotrophy in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 2014, 80, 2094–2101. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Boller, T.; Wiemken, A. Trehalose becomes the most abundant non-structural carbohydrate during senescence of soybean nodules. J. Exp. Bot. 2001, 52, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Streeter, J.G. Effect of trehalose on survival of Bradyrhizobium japonicum during desiccation. J. Appl. Microbiol. 2003, 95, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Streeter, J.G.; Gomez, M.L. Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and in bacteroids from soybean nodules. Appl. Environ. Microbiol. 2006, 72, 4250–4255. [Google Scholar] [CrossRef] [PubMed]
- Cytryn, E.J.; Sangurdekar, D.P.; Streeter, J.G.; Franck, W.L.; Chang, W.S.; Stacey, G.; Emerich, D.W.; Joshi, T.; Xu, D.; Sadowsky, M.J. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 2007, 189, 6751–6762. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, M.; Cytryn, E.J.; Sadowsky, M.J. Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation. Appl. Environ. Microbiol. 2010, 76, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.; Meyer, L.; Studer, D.; Regensburger, B.; Hennecke, H. Insertion and deletion mutations within the nif region of Rhizobium japonicum. Plant Mol. Biol. 1984, 3, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.M.; Alvarez-Morales, A.; Hennecke, H. The pleiotropic nature of symbiotic regulatory mutants: Bradyrhizobium japonicum nifA gene is involved in control of nif gene expression and formation of determinate symbiosis. EMBO J. 1986, 5, 1165–1173. [Google Scholar] [PubMed]
- Cermola, M.; Fedorova, E.; Tate, R.; Riccio, A.; Favre, R.; Patriarca, E.J. Nodule invasion and symbiosome differentiation during Rhizobium etli–Phaseolus vulgaris symbiosis. Mol. Plant Microbe Interact. 2000, 13, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Studer, D.; Gloudemans, T.; Franssen, H.J.; Fischer, H.M.; Bisseling, T.; Hennecke, H. Involvement of the bacterial nitrogen-fixation regulatory gene nifA in control of nodule-specific host-plant gene-expression. Eur. J. Cell Biol. 1987, 45, 177–184. [Google Scholar]
- Cabrera, J.J.; Salas, A.; Torres, M.J.; Bedmar, E.J.; Richardson, D.J.; Gates, A.J.; Delgado, M.J. An integrated biochemical system for nitrate assimilation and nitric oxide detoxification in Bradyrhizobium japonicum. Biochem. J. 2016, 473, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Boscari, A.; Castella, C.; Rovere, M.; Puppo, A.; Brouquisse, R. Nitric oxide: A multifaceted regulator of the nitrogen-fixing symbiosis. J. Exp. Bot. 2015, 66, 2877–2887. [Google Scholar] [CrossRef] [PubMed]
- Baudouin, E.; Pieuchot, L.; Engler, G.; Pauly, N.; Puppo, A. Nitric oxide is formed in Medicago truncatula–Sinorhizobium meliloti functional nodules. Mol. Plant Microbe Interact. 2006, 19, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M.; Fischer, H.M.; Hennecke, H.; Werner, D. Accumulation of the phytoalexin glyceollin I in soybean nodules infected by a Bradyrhizobium japonicum nifA mutant. Z. Naturforschung C 1991, 46, 318–320. [Google Scholar]
- Krause, A.; Doerfel, A.; Göttfert, M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2002, 15, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef] [PubMed]
- Deakin, W.J.; Broughton, W.J. Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 2009, 7, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Xin, D.W.; Liao, S.; Xie, Z.P.; Hann, D.R.; Steinle, L.; Boller, T.; Staehelin, C. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 2012, 8, e1002707. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Guerrero, I.; Pérez-Montaňo, F.; Monreal, J.A.; Preston, G.M.; Fones, H.; Vioque, B.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 type 3 secretion system suppresses early defense responses to effectively nodulate soybean. Mol. Plant Microbe Interact. 2015, 28, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Tsukui, T.; Eda, S.; Kaneko, T.; Sato, S.; Okazaki, S.; Kakizaki-Chiba, K.; Itakura, M.; Mitsui, H.; Yamashita, A.; Terasawa, K.; et al. The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microb. 2013, 79, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Minamisawa, K.; Uchiumi, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Iriguchi, M.; Kawashima, K.; et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.; Hennecke, H. Localized mutagenesis in Rhizobium japonicum. Mol. Gen. Genet. 1984, 193, 46–52. [Google Scholar] [CrossRef]
- Regensburger, B.; Hennecke, H. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 1983, 135, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Qeli, E.; Ahrens, C.H. PeptideClassifier for protein inference and targeted quantitative proteomics. Nat. Biotechnol. 2010, 28, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Gerster, S.; Qeli, E.; Ahrens, C.H.; Buhlmann, P. Protein and gene model inference based on statistical modeling in k-partite graphs. Proc. Natl. Acad. Sci. USA 2010, 107, 12101–12106. [Google Scholar] [CrossRef] [PubMed]
- Omasits, U.; Quebatte, M.; Stekhoven, D.J.; Fortes, C.; Roschitzki, B.; Robinson, M.D.; Dehio, C.; Ahrens, C.H. Directed shotgun proteomics guided by saturated RNA-seq identifies a complete expressed prokaryotic proteome. Genome Res. 2013, 23, 1916–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Čuklina, J.; Hahn, J.; Imakaev, M.; Omasits, U.; Förstner, K.U.; Ljubimov, N.; Goebel, M.; Pessi, G.; Fischer, H.M.; Ahrens, C.H.; et al. Genome-wide transcription start site mapping of Bradyrhizobium japonicum grown free-living or in symbiosis—A rich resource to identify new transcripts, proteins and to study gene regulation. BMC Genom. 2016, 17, 302. [Google Scholar] [CrossRef] [PubMed]


| Strain a | Host Plant | Number of Biological Replicates | dpi a | ||||
|---|---|---|---|---|---|---|---|
| WT | soybean | 3 | 13 | ||||
| WT | soybean | 3 | 21 | ||||
| WT | soybean | 3 | 31 | ||||
| A9 (nifA mutant) | soybean | 3 | 21 | ||||
| H1 (nifH mutant) | soybean | 3 | 21 | ||||
| none (uninfected roots) | soybean | 3 | 21 | ||||
| WT | cowpea | 3 | 21 | ||||
| none (uninfected roots) | cowpea | 3 | 21 | ||||
| WT | mungbean | 3 | 21 | ||||
| none (uninfected roots) | mungbean | 3 | 21 | ||||
| WT | siratro | 3 | 31 | ||||
| none (uninfected roots) | siratro | 2 | 31 | ||||
| Experimental Comparison a | # incr met b | # decr met c | Table | ||||
| 2.2. Host-Specific Nodule and Root Metabolome (4 host plants) | |||||||
| WT vs. none (uninfected roots) | 132 | 21 | Table S2 | ||||
| WT or none (soybean) vs. WT or none (cowpea, mungbean, siratro) | 67 | nd | Table S3 | ||||
| WT or none (mungbean) vs. WT or none (cowpea, soybean, siratro) | 30 | nd | Table S3 | ||||
| WT or none (siratro) vs. WT or none (cowpea, mungbean, soybean) | 17 | nd | Table S3 | ||||
| WT or none (cowpea) vs. WT or none (mungbean, soybean, siratro) | 17 | nd | Table S3 | ||||
| 2.3. Metabolome of different stages of nodule development (soybean) | |||||||
| WT 13 dpi vs. 21 and 31 dpi | 6 | nd | Table 2 | ||||
| WT 21 dpi vs. 13 and 31 dpi | 1 | nd | Table 2 | ||||
| WT 31 dpi vs. 13 and 21 dpi | 4 | nd | Table 2 | ||||
| 2.4. Metabolome of nodules induced by a nifA and nifH mutant (soybean) | |||||||
| WT vs. A9 (nifA mutant) | 25 | 112 | |||||
| WT vs. H1 (nifH mutant) | 19 | 69 | |||||
| Metabolites Specific for the Indicated Time Point a | ID a | log2 13 vs 21 b | log2 13 vs 31 b | log2 21 vs 31 b |
|---|---|---|---|---|
| 13 dpi | ||||
| Tryptophan | C00078 | 2.4 | 2.0 | nr |
| Cyclohexylformamide | C11519 | 0.9 | 1.2 | nr |
| Glutamyl-taurine | C05844 | 0.9 | 0.9 | nr |
| Oxalate | C00209 | 0.6 | 0.7 | nr |
| Fumarate | C00122 | 0.6 | 1.3 | ns |
| Phosphoenolpyruvate | C00074 | 0.5 | 0.8 | nr |
| 21 dpi | ||||
| Trehalose 6-phosphate | C00689 | −1.2 | −0.3 | 0.9 |
| 31 dpi | ||||
| Glucosamine 6-phosphate | C00352 | nr | −1.2 | −0.8 |
| Indole-3-acetate | C00954 | nr | −1.0 | −0.7 |
| Isopropylmaleate | C02631 | nr | −0.8 | −0.8 |
| AMP | C00020 | nr | −0.7 | −0.7 |
| ORF No. a | Description b | Gene Name b | log2(nifA vs. wt) c | log2(nifH vs. wt) c |
|---|---|---|---|---|
| Energy production and conversion | ||||
| bll1718 | C4-dicarboxylate transport protein | dctA | −4.6 | −2.0 |
| bll2063 | phenolhydroxylase homolog | nrgC | −6.2 | |
| bll4571 | putative ferredoxin—nitrite reductase | 2.2 | 5.0 | |
| bll6940 | HupC protein | hupC | −5.4 | |
| bll6941 | uptake hydrogenase large subunit | hupL | −4.1 | |
| bll6942 | uptake hydrogenase precursor | hupA | −6.6 | |
| blr1721 | uptake hydrogenase large subunit homolog | hupL | −4.2 | |
| blr1724 | HupD protein homolog | −4.3 | ||
| blr1743 | nitrogenase molybdenum-iron protein alpha chain | nifD | −5.4 | |
| blr1744 | nitrogenase molybdenum-iron protein beta chain | nifK | −5.2 | |
| blr1745 | nitrogenase molybdenum-cofactor synthesis protein | nifE | −6.0 | |
| blr1746 | nitrogenase molybdenum-cofactor synthesis protein | nifN | −5.6 | |
| blr1765 | Ferredoxin | fer2 | −4.3 | |
| blr1773 | electron transfer flavoprotein alpha chain | fixB | −5.0 | −1.2 |
| blr1774 | Flavoprotein | fixC | −5.2 | |
| blr1816 | RhcN protein | rhcN | 4.4 | |
| blr1853 | cytochrome P450 family protein | −6.0 | ||
| blr2038 | electron transfer flavoprotein beta chain | fixA | −4.6 | |
| blr2143 | similar to cytochrome P450-family protein | −4.9 | ||
| blr3719 | hypothetical protein | −3.6 | −4.3 | |
| blr3722 | dihydrolipoamide dehydrogenase | lpd | −2.8 | −4.9 |
| bsr1739 | Ferredoxin | fdxN | −5.1 | |
| bsr1760 | ferredoxin-like protein | frxA | −5.9 | |
| bsr1775 | probable ferredoxin | fixX | −6.3 | |
| Amino acid transport and metabolism | ||||
| blr1756 | nitrogenase metalloclusters biosynthesis protein | nifS | −5.4 | |
| blr1971 | putative peptidase | −4.6 | ||
| blr2071 | similar to inosamine-phosphate amidinotransferase | -4.9 | ||
| blr2106 | l-ectoine synthase | ectC | −6.5 | |
| blr2136 | putative aminotransferase | −5.4 | ||
| Carbohydrate transport and metabolism | ||||
| blr1656 | putative glycosyl hydrolase | 4.8 | ||
| blr2581 | putative D-fructose-1,6-bisphosphatase protein | cbbF | 5.0 | |
| Coenzyme transport and metabolism | ||||
| blr1686 | putative aminotransferase protein | −6.0 | ||
| blr1852 | similar to pantoate—β-alanine ligase | −4.3 | ||
| Translation, ribosomal structure and biogenesis | ||||
| blr2135 | hypothetical protein | −5.0 | ||
| Transcription | ||||
| bll1906 | N-acetyltransferase NrgA homolog | −5.0 | ||
| blr1880 | transcriptional regulatory protein LuxR family | −5.0 | ||
| Replication and repair | ||||
| blr8234 | unknown protein | −4.7 | ||
| Cell wall/membrane/envelop biogenesis | ||||
| bll1872 | hypothetical protein | −5.5 | ||
| bll1944 | hypothetical protein | −5.4 | ||
| bll2085 | hypothetical protein | −4.3 | ||
| Post-translational modification, protein turnover, and chaperones | ||||
| bll1777 | alkyl hydroperoxide reductase | ahpC | −6.0 | |
| bll2059 | GroEL3 chaperonin | groEL3 | −4.8 | |
| bll2060 | GroES3 chaperonin | groES3 | −4.9 | |
| blr1879 | hypothetical protein | −4.4 | ||
| Inorganic ion transport and metabolism | ||||
| bll2801 | probable potential formate transporter | 4.2 | ||
| bll4570 | probable sulfite reductase (NADPH) flavoprotein | 1.6 | 4.8 | |
| bll5736 | putative thiosulfate sulfurtransferase precursor | 4.0 | 5.8 | |
| blr1719 | molybdenum transport system permease protein | modB | −4.3 | |
| blr1769 | dinitrogenase reductase protein | nifH | −5.3 | −3.3 |
| blr2803 | ABC transporter nitrate-binding protein | nrtA | 1.6 | 4.2 |
| blr3278 | hypothetical protein | −4.1 | ||
| blr6951 | molybdenum ABC transporter Molybdate-binding protein | modA | −4.9 | |
| blr7315 | unknown protein | 4.3 | ||
| Secondary metabolites biosynthesis, transport, and catabolism | ||||
| bll2125 | probable dioxygenase | −5.0 | ||
| blr2036 | Oxidoreductase | fixR | −4.3 | |
| blr2108 | probable peptide synthetase | −4.6 | ||
| blr2131 | probable oxygenase | −6.0 | ||
| blr2133 | hypothetical protein | −5.3 | ||
| blr2144 | cytochrome P-450 BJ-1 | cyp112 | −4.5 | |
| blr2145 | cytochrome P-450 BJ-3 | cyp114 | −4.2 | |
| bsr1757 | nitrogen fixation protein | −5.0 | ||
| General functional prediction only | ||||
| bll1776 | alkyl hydroperoxide reductase | ahpD | −4.2 | |
| blr1759 | FeMo cofactor biosynthesis protein | nifB | −5.1 | |
| blr2041 | unknown protein | −4.2 | ||
| blr2042 | hypothetical protein | −4.8 | ||
| blr7556 | non-heme haloperoxidase | −1.9 | 4.4 | |
| Function unknown | ||||
| bll1754 | hypothetical protein | −4.8 | ||
| bll1767 | hypothetical protein | −6.0 | ||
| bll1810 | hypothetical protein | 5.4 | ||
| bll1979 | hypothetical protein | −5.6 | ||
| bll1980 | hypothetical protein | −4.8 | ||
| bll1981 | hypothetical protein | −4.9 | ||
| bll2003 | unknown protein | −4.3 | ||
| bll2009 | hypothetical protein | −5.6 | ||
| bll4177 | hypothetical protein | 5.5 | ||
| bll5738 | unknown protein | 4.8 | 6.8 | |
| bll6381 | unknown protein | −4.3 | −1.0 | |
| bll6552 | hypothetical protein | 4.5 | ||
| blr1649 | unknown protein | 5.3 | ||
| blr1676 | hypothetical protein | 4.2 | ||
| blr1704 | hypothetical protein | 6.1 | ||
| blr1705 | unknown protein | 4.6 | ||
| blr1747 | iron-molibdenum cofactor processing protein | nifX | −4.6 | |
| blr1748 | hypothetical protein | −6.3 | ||
| blr1755 | R. etli iscN homolog | −4.7 | ||
| blr1761 | iron-sulfur cofactor synthesis protein | nifZ | −4.6 | |
| blr1770 | molybdenum processing protein | nifQ | −5.1 | −4.5 |
| blr1771 | nitrogenase stabilizing-protective protein | nifW | −5.7 | −3.9 |
| blr1806 | unknown protein | 4.9 | ||
| blr1812 | nodulation protein | nolB | 5.9 | |
| blr1814 | nodulation protein | nolU | 5.0 | |
| blr1817 | hypothetical protein | 5.1 | ||
| blr1851 | unknown protein | −4.7 | ||
| blr2132 | unknown protein | −6.7 | ||
| blr2140 | hypothetical protein | 4.2 | ||
| blr2505 | hypothetical protein | −4.3 | ||
| blr6172 | hypothetical protein | 4.7 | ||
| blr7321 | hypothetical protein | 4.3 | ||
| blr7327 | hypothetical protein | 6.0 | ||
| bsr1749 | hypothetical protein | −4.4 | ||
| Intracellular trafficking, secretion, and vesicular transport | ||||
| blr1813 | RhcJ protein | rhcJ | 5.7 | |
| blr1819 | RhcR protein | rhcR | 4.5 | |
| bsr1820 | RhcS protein | rhcS | 4.9 | |
| Other | ||||
| bll1634 | unknown protein | −4.9 | ||
| bll1636 | unknown protein | −4.4 | ||
| bll1801 | hypothetical protein | 4.3 | ||
| bll1804 | unknown protein | 5.4 | ||
| bll1846 | unknown protein | 4.3 | ||
| bll1858 | hypothetical protein | −5.2 | ||
| bll1877 | unknown protein | 4.4 | ||
| bll2004 | unknown protein | −4.9 | ||
| bll2067 | nodulate formation efficiency C protein | nfeC | −4.9 | |
| bll6154 | unknown protein | 3.1 | 4.9 | |
| blr1638 | unknown protein | −5.4 | ||
| blr1650 | unknown protein | 4.9 | ||
| blr1726 | unknown protein | −7.0 | ||
| blr1728 | HupK protein homolog | hupK | −4.5 | |
| blr1763 | unknown protein | −6.1 | ||
| blr1850 | unknown protein | −5.2 | ||
| blr1954 | unknown protein | −6.7 | ||
| blr1964 | putative sugar hydrolase | −5.4 | ||
| blr1992 | unknown protein | −5.5 | ||
| blr2011 | unknown protein | −4.9 | ||
| blr2069 | unknown protein | −4.4 | ||
| blr2134 | hypothetical protein | −5.1 | ||
| blr7314 | unknown protein | 5.6 | ||
| bsl1637 | unknown protein | −5.7 | ||
| bsl1651 | unknown protein | 4.2 | ||
| bsl1652 | unknown protein | 4.8 | ||
| bsl1808 | unknown protein | 5.1 | ||
| bsl1857 | unknown protein | −4.8 | ||
| bsl2596 | unknown protein | 5.0 | ||
| bsr1758 | unknown protein | −5.1 | ||
| bsr1764 | unknown protein | −4.4 | ||
| bsr1907 | unknown protein | −4.0 | ||
| bsr2005 | unknown protein | −4.4 | ||
| bsr2010 | unknown protein | −6.0 | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lardi, M.; Murset, V.; Fischer, H.-M.; Mesa, S.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures. Int. J. Mol. Sci. 2016, 17, 815. https://doi.org/10.3390/ijms17060815
Lardi M, Murset V, Fischer H-M, Mesa S, Ahrens CH, Zamboni N, Pessi G. Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures. International Journal of Molecular Sciences. 2016; 17(6):815. https://doi.org/10.3390/ijms17060815
Chicago/Turabian StyleLardi, Martina, Valérie Murset, Hans-Martin Fischer, Socorro Mesa, Christian H. Ahrens, Nicola Zamboni, and Gabriella Pessi. 2016. "Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures" International Journal of Molecular Sciences 17, no. 6: 815. https://doi.org/10.3390/ijms17060815
APA StyleLardi, M., Murset, V., Fischer, H.-M., Mesa, S., Ahrens, C. H., Zamboni, N., & Pessi, G. (2016). Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures. International Journal of Molecular Sciences, 17(6), 815. https://doi.org/10.3390/ijms17060815