Reconstruction and Application of Protein–Protein Interaction Network
Abstract
:1. Introduction
2. PINs for Various Organisms
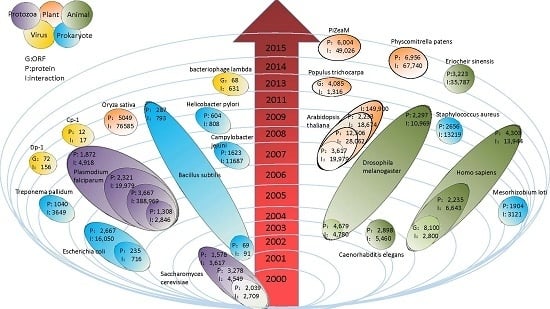
2.1. Prokaryote
2.2. Eukaryote
2.2.1. Protozoa
2.2.2. Plants
2.2.3. Animals
2.3. Virus
3. Major Techniques Used in PIN Reconstruction
3.1. Yeast Two-Hybrid (Y2H)
3.2. Affinity Purification and Mass Spectrometry
3.3. Prediction Based on Computational Method
3.3.1. Interolog-Based Method
3.3.2. Prediction Based on Genetic Algorithms
4. Application
4.1. Function Annotation of Proteins
4.1.1. Annotation Based on Adjacency Proteins
4.1.2. Annotation Based on Cluster Analysis
4.2. Subsystem Investigation
4.3. Evolutionary Analysis
4.4. Hub Protein Analysis
4.5. Regulation Mechanism Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Snider, J.; Kotlyar, M.; Saraon, P.; Yao, Z.; Jurisica, I.; Stagljar, I. Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 2015, 11, 848. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Biological robustness. Nat. Rev. Genet. 2004, 5, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.; Lachaize, C.; Janody, F.; Bellon, B.; Roder, L.; Euzenat, J.; Rechenmann, F.; Jacq, B. Grasping at molecular interactions and genetic networks in drosophila melanogaster using flynets, an internet database. Nucleic Acids Res. 1999, 27, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Mosca, R.; Pons, T.; Ceol, A.; Valencia, A.; Aloy, P. Towards a detailed atlas of protein-protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K.; Kim, H.B. Protein interaction network related to helicobacter pylori infection response. World J. Gastroenterol. 2009, 15, 4518–4528. [Google Scholar] [CrossRef] [PubMed]
- Geisler-Lee, J.; O’Toole, N.; Ammar, R.; Provart, N.J.; Millar, A.H.; Geisler, M. A predicted interactome for arabidopsis. Plant Physiol. 2007, 145, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Schwikowski, B.; Uetz, P.; Fields, S. A network of protein-protein interactions in yeast. Nat. Biotechnol. 2000, 18, 1257–1261. [Google Scholar] [CrossRef] [PubMed]
- Rual, J.F.; Venkatesan, K.; Hao, T.; Hirozane-Kishikawa, T.; Dricot, A.; Li, N.; Berriz, G.F.; Gibbons, F.D.; Dreze, M.; Ayivi-Guedehoussou, N.; et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005, 437, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Butland, G.; Peregrin-Alvarez, J.M.; Li, J.; Yang, W.; Yang, X.; Canadien, V.; Starostine, A.; Richards, D.; Beattie, B.; Krogan, N.; et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 2005, 433, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Arifuzzaman, M.; Maeda, M.; Itoh, A.; Nishikata, K.; Takita, C.; Saito, R.; Ara, T.; Nakahigashi, K.; Huang, H.C.; Hirai, A.; et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006, 16, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Greenbaum, D.; Xin Lu, H.; Zhu, X.; Gerstein, M. Genomic analysis of essentiality within protein networks. Trends Genet. TIG 2004, 20, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Noirot-Gros, M.F.; Dervyn, E.; Wu, L.J.; Mervelet, P.; Errington, J.; Ehrlich, S.D.; Noirot, P. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. USA 2002, 99, 8342–8347. [Google Scholar] [CrossRef] [PubMed]
- Marchadier, E.; Carballido-Lopez, R.; Brinster, S.; Fabret, C.; Mervelet, P.; Bessieres, P.; Noirot-Gros, M.F.; Fromion, V.; Noirot, P. An expanded protein-protein interaction network in bacillus subtilis reveals a group of hubs: Exploration by an integrative approach. Proteomics 2011, 11, 2981–2991. [Google Scholar] [CrossRef] [PubMed]
- Magrane, M.; Consortium, U. Uniprot knowledgebase: A hub of integrated protein data. Database J. Biol. Databases Curation 2011. [Google Scholar] [CrossRef] [PubMed]
- McDowall, M.D.; Scott, M.S.; Barton, G.J. Pips: Human protein-protein interaction prediction database. Nucleic Acids Res. 2009, 37, D651–D656. [Google Scholar] [CrossRef] [PubMed]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Parrish, J.R.; Yu, J.; Liu, G.; Hines, J.A.; Chan, J.E.; Mangiola, B.A.; Zhang, H.; Pacifico, S.; Fotouhi, F.; DiRita, V.J.; et al. A proteome-wide protein interaction map for campylobacter jejuni. Genome Biol. 2007, 8, R130. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Rajagopala, S.V.; Goll, J.; Hauser, R.; McKevitt, M.T.; Palzkill, T.; Uetz, P. The binary protein interactome of treponema pallidum—The syphilis spirochete. PLoS ONE 2008, 3, e2292. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P.; Hughes, R.E. Systematic and large-scale two-hybrid screens. Curr. Opin. Microbiol. 2000, 3, 303–308. [Google Scholar] [CrossRef]
- Uetz, P.; Giot, L.; Cagney, G.; Mansfield, T.A.; Judson, R.S.; Knight, J.R.; Lockshon, D.; Narayan, V.; Srinivasan, M.; Pochart, P.; et al. A comprehensive analysis of protein-protein interactions in saccharomyces cerevisiae. Nature 2000, 403, 623–627. [Google Scholar] [PubMed]
- Ito, T.; Tashiro, K.; Muta, S.; Ozawa, R.; Chiba, T.; Nishizawa, M.; Yamamoto, K.; Kuhara, S.; Sakaki, Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 1143–1147. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Chiba, T.; Ozawa, R.; Yoshida, M.; Hattori, M.; Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 2001, 98, 4569–4574. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Gruhler, A.; Heilbut, A.; Bader, G.D.; Moore, L.; Adams, S.L.; Millar, A.; Taylor, P.; Bennett, K.; Boutilier, K.; et al. Systematic identification of protein complexes in saccharomyces cerevisiae by mass spectrometry. Nature 2002, 415, 180–183. [Google Scholar] [CrossRef] [PubMed]
- LaCount, D.J.; Vignali, M.; Chettier, R.; Phansalkar, A.; Bell, R.; Hesselberth, J.R.; Schoenfeld, L.W.; Ota, I.; Sahasrabudhe, S.; Kurschner, C.; et al. A protein interaction network of the malaria parasite plasmodium falciparum. Nature 2005, 438, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Wuchty, S.; Adams, J.H.; Ferdig, M.T. A comprehensive plasmodium falciparum protein interaction map reveals a distinct architecture of a core interactome. Proteomics 2009, 9, 1841–1849. [Google Scholar] [CrossRef] [PubMed]
- Date, S.V.; Stoeckert, C.J., Jr. Computational modeling of the plasmodium falciparum interactome reveals protein function on a genome-wide scale. Genome Res. 2006, 16, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Wuchty, S.; Ipsaro, J.J. A draft of protein interactions in the malaria parasite P. falciparum. J. Proteome Res. 2007, 6, 1461–1470. [Google Scholar] [CrossRef] [PubMed]
- Mitrofanova, A.; Kleinberg, S.; Carlton, J.; Kasif, S.; Mishra, B. Predicting malaria interactome classifications from time-course transcriptomic data along the intraerythrocytic developmental cycle. Artif. Intell. Med. 2010, 49, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Musungu, B.; Bhatnagar, D.; Brown, R.L.; Fakhoury, A.M.; Geisler, M. A predicted protein interactome identifies conserved global networks and disease resistance subnetworks in maize. Front. Genet. 2015, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Schuette, S.; Piatkowski, B.; Corley, A.; Lang, D.; Geisler, M. Predicted protein-protein interactions in the moss physcomitrella patens: A new bioinformatic resource. BMC Bioinform. 2015, 16, 89. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, P.; Li, G.; Xu, F.; Zhao, C.; Li, Y.; Yang, Z.; Wang, G.; Yu, Q.; Shi, T. Atpid: Arabidopsis thaliana protein interactome database—An integrative platform for plant systems biology. Nucleic Acids Res. 2008, 36, D999–D1008. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zang, W.; Li, Y.; Xu, F.; Wang, J.; Shi, T. Atpid: The overall hierarchical functional protein interaction network interface and analytic platform for arabidopsis. Nucleic Acids Res. 2011, 39, D1130–D1133. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Kegg as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- De Bodt, S.; Proost, S.; van de Poele, K.; Rouze, P.; van de Peer, Y. Predicting protein-protein interactions in arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC Genom. 2009, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Shen, X.; Chen, X. Pair: The predicted arabidopsis interactome resource. Nucleic Acids Res. 2011, 39, D1134–D1140. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zhou, X.; Shen, X.; Mao, C.; Chen, X. The predicted arabidopsis interactome resource and network topology-based systems biology analyses. Plant Cell 2011, 23, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Rodgers-Melnick, E.; Culp, M.; DiFazio, S.P. Predicting whole genome protein interaction networks from primary sequence data in model and non-model organisms using ents. BMC Genom. 2013, 14, 608. [Google Scholar] [CrossRef] [PubMed]
- Yellaboina, S.; Tasneem, A.; Zaykin, D.V.; Raghavachari, B.; Jothi, R. Domine: A comprehensive collection of known and predicted domain-domain interactions. Nucleic Acids Res. 2011, 39, D730–D735. [Google Scholar] [CrossRef] [PubMed]
- Giot, L.; Bader, J.S.; Brouwer, C.; Chaudhuri, A.; Kuang, B.; Li, Y.; Hao, Y.L.; Ooi, C.E.; Godwin, B.; Vitols, E.; et al. A protein interaction map of drosophila melanogaster. Science 2003, 302, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Rual, J.F.; Zhai, B.; Mintseris, J.; Vaidya, P.; Vaidya, N.; Beekman, C.; Wong, C.; Rhee, D.Y.; Cenaj, O.; et al. A protein complex network of drosophila melanogaster. Cell 2011, 147, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Armstrong, C.M.; Bertin, N.; Ge, H.; Milstein, S.; Boxem, M.; Vidalain, P.O.; Han, J.D.; Chesneau, A.; Hao, T.; et al. A map of the interactome network of the metazoan C. elegans. Science 2004, 303, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Ewing, R.M.; Chu, P.; Elisma, F.; Li, H.; Taylor, P.; Climie, S.; McBroom-Cerajewski, L.; Robinson, M.D.; O’Connor, L.; Li, M.; et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Tasan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A proteome-scale map of the human interactome network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Zeng, Z.; Wang, B.; Zhang, Y.; Liu, Y.; Geng, X.; Sun, J. The protein-protein interaction network of eyestalk, Y-organ and hepatopancreas in chinese mitten CRAB Eriocheir sinensis. BMC Syst. Biol. 2014, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Hauser, R.; Blasche, S.; Dokland, T.; Haggard-Ljungquist, E.; von Brunn, A.; Salas, M.; Casjens, S.; Molineux, I.; Uetz, P. Bacteriophage protein-protein interactions. Adv. Virus Res. 2012, 83, 219–298. [Google Scholar] [PubMed]
- Court, D.L.; Oppenheim, A.B.; Adhya, S.L. A new look at bacteriophage lambda genetic networks. J. Bacteriol. 2007, 189, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in bacteriophage lambda development. Ann. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef] [PubMed]
- Rajagopala, S.V.; Casjens, S.; Uetz, P. The protein interaction map of bacteriophage lambda. BMC Microbiol. 2011, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Blasche, S.; Wuchty, S.; Rajagopala, S.V.; Uetz, P. The protein interaction network of bacteriophage lambda with its host, Escherichia coli. J. Virol. 2013, 87, 12745–12755. [Google Scholar] [CrossRef] [PubMed]
- De Chassey, B.; Navratil, V.; Tafforeau, L.; Hiet, M.S.; Aublin-Gex, A.; Agaugue, S.; Meiffren, G.; Pradezynski, F.; Faria, B.F.; Chantier, T.; et al. Hepatitis c virus infection protein network. Mol. Syst. Biol. 2008, 4, 230. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, M.A.; Venkatesan, K.; Xing, L.; Chase, M.R.; Vazquez, A.; Holthaus, A.M.; Ewence, A.E.; Li, N.; Hirozane-Kishikawa, T.; Hill, D.E.; et al. Epstein-barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. USA 2007, 104, 7606–7611. [Google Scholar] [CrossRef] [PubMed]
- Dyer, M.D.; Murali, T.M.; Sobral, B.W. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008, 4, e32. [Google Scholar] [CrossRef] [PubMed]
- Navratil, V.; de Chassey, B.; Meyniel, L.; Pradezynski, F.; Andre, P.; Rabourdin-Combe, C.; Lotteau, V. System-level comparison of protein-protein interactions between viruses and the human type I interferon system network. J. Proteome Res. 2010, 9, 3527–3536. [Google Scholar] [CrossRef] [PubMed]
- Fields, S.; Song, O. A novel genetic system to detect protein-protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, A.; Colas, P. Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol. Sci. 2012, 33, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.; Trabalzini, L. The yeast two-hybrid and related methods as powerful tools to study plant cell signalling. Plant Mol. Biol. 2013, 83, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Stasi, M.; de Luca, M.; Bucci, C. Two-hybrid-based systems: Powerful tools for investigation of membrane traffic machineries. J. Biotechnol. 2015, 202, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Tavernier, J.; Eyckerman, S.; Lemmens, I.; Van der Heyden, J.; Vandekerckhove, J.; van Ostade, X. Mappit: A cytokine receptor-based two-hybrid method in mammalian cells. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2002, 32, 1397–1404. [Google Scholar] [CrossRef]
- Rigaut, G.; Shevchenko, A.; Rutz, B.; Wilm, M.; Mann, M.; Seraphin, B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999, 17, 1030–1032. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.C.; Aloy, P.; Grandi, P.; Krause, R.; Boesche, M.; Marzioch, M.; Rau, C.; Jensen, L.J.; Bastuck, S.; Dumpelfeld, B.; et al. Proteome survey reveals modularity of the yeast cell machinery. Nature 2006, 440, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Janga, S.C.; Babu, M.; Diaz-Mejia, J.J.; Butland, G.; Yang, W.; Pogoutse, O.; Guo, X.; Phanse, S.; Wong, P.; et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009, 7, e96. [Google Scholar] [CrossRef] [PubMed]
- Kuhner, S.; van Noort, V.; Betts, M.J.; Leo-Macias, A.; Batisse, C.; Rode, M.; Yamada, T.; Maier, T.; Bader, S.; Beltran-Alvarez, P.; et al. Proteome organization in a genome-reduced bacterium. Science 2009, 326, 1235–1240. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, K.; Ma, B.; Nussinov, R. Is allostery an intrinsic property of all dynamic proteins? Proteins 2004, 57, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Dreze, M.; Carvunis, A.R.; Charloteaux, B.; Galli, M.; Pevzner, S.J.; Tasan, M.; Ahn, Y.-Y.; Balumuri, P.; Barabási, A.-L.; Bautista, V.; et al. Evidence for network evolution in an arabidopsis interactome map. Science 2011, 333, 601–607. [Google Scholar]
- Hirsh, E.; Sharan, R. Identification of conserved protein complexes based on a model of protein network evolution. Bioinformatics 2007, 23, e170–e176. [Google Scholar] [CrossRef] [PubMed]
- De Smet, R.; van de Peer, Y. Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr. Opin. Plant Biol. 2012, 15, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Matthews, L.R.; Vaglio, P.; Reboul, J.; Ge, H.; Davis, B.P.; Garrels, J.; Vincent, S.; Vidal, M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001, 11, 2120–2126. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef] [PubMed]
- Teichmann, S.A.; Rison, S.C.; Thornton, J.M.; Riley, M.; Gough, J.; Chothia, C. The evolution and structural anatomy of the small molecule metabolic pathways in escherichia coli. J. Mol. Biol. 2001, 311, 693–708. [Google Scholar] [CrossRef] [PubMed]
- Faure, G.; Andreani, J.; Guerois, R. Interevol database: Exploring the structure and evolution of protein complex interfaces. Nucleic Acids Res. 2012, 40, D847–D856. [Google Scholar] [CrossRef] [PubMed]
- Fariselli, P.; Pazos, F.; Valencia, A.; Casadio, R. Prediction of protein-protein interaction sites in heterocomplexes with neural networks. Eur. J. Biochem./FEBS 2002, 269, 1356–1361. [Google Scholar] [CrossRef]
- Nguyen, T.P.; Ho, T.B. An integrative domain-based approach to predicting protein-protein interactions. J. Bioinform. Comput. Biol. 2008, 6, 1115–1132. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.A.; Li, K.; Kanade, T. A semi-markov model for mitosis segmentation in time-lapse phase contrast microscopy image sequences of stem cell populations. IEEE Trans. Med. Imaging 2012, 31, 359–369. [Google Scholar] [PubMed]
- Brunk, E.; Mih, N.; Monk, J.; Zhang, Z.; O’Brien, E.J.; Bliven, S.E.; Chen, K.; Chang, R.L.; Bourne, P.E.; Palsson, B.O. Systems biology of the structural proteome. BMC Syst. Biol. 2016, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, Y.; Vakser, I.A. Dockground protein-protein docking decoy set. Bioinformatics 2008, 24, 2634–2635. [Google Scholar] [CrossRef] [PubMed]
- Kundrotas, P.J.; Zhu, Z.; Vakser, I.A. Gwidd: Genome-wide protein docking database. Nucleic Acids Res. 2010, 38, D513–D517. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Kerrien, S.; Abbani, S.; Aranda, B.; Bhate, J.; Bidwell, S.; Bridge, A.; Briganti, L.; Brinkman, F.S.; Cesareni, G.; et al. Protein interaction data curation: The international molecular exchange (imex) consortium. Nat. Methods 2012, 9, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Alfarano, C.; Andrade, C.E.; Anthony, K.; Bahroos, N.; Bajec, M.; Bantoft, K.; Betel, D.; Bobechko, B.; Boutilier, K.; Burgess, E.; et al. The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 2005, 33, D418–D424. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Breitkreutz, B.J.; Oughtred, R.; Boucher, L.; Heinicke, S.; Chen, D.; Stark, C.; Breitkreutz, A.; Kolas, N.; O’Donnell, L.; et al. The biogrid interaction database: 2015 update. Nucleic Acids Res. 2015, 43, D470–D478. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Wakamatsu, A.; Kawamura, Y.; Kimura, K.; Yamamoto, J.; Nishikawa, T.; Kisu, Y.; Sugano, S.; Goshima, N.; Isogai, T.; et al. Human gene and protein database (HGPD): A novel database presenting a large quantity of experiment-based results in human proteomics. Nucleic Acids Res. 2009, 37, D762–D766. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.R.; Jurisica, I. Online predicted human interaction database. Bioinformatics 2005, 21, 2076–2082. [Google Scholar] [CrossRef] [PubMed]
- Lopez, Y.; Nakai, K.; Patil, A. Hitpredict version 4: Comprehensive reliability scoring of physical protein-protein interactions from more than 100 species. Database J. Biol. Databases Curation 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, M.; Pastrello, C.; Sheahan, N.; Jurisica, I. Integrated interactions database: Tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2016, 44, D536–D541. [Google Scholar] [CrossRef] [PubMed]
- Aranda, B.; Achuthan, P.; Alam-Faruque, Y.; Armean, I.; Bridge, A.; Derow, C.; Feuermann, M.; Ghanbarian, A.T.; Kerrien, S.; Khadake, J.; et al. The intact molecular interaction database in 2010. Nucleic Acids Res. 2010, 38, D525–D531. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.; Razick, S.; Turinsky, A.L.; Vlasblom, J.; Crowdy, E.K.; Cho, E.; Morrison, K.; Donaldson, I.M.; Wodak, S.J. Irefweb: Interactive analysis of consolidated protein interaction data and their supporting evidence. Database J. Biol. Databases Curation 2010, 2010, baq023. [Google Scholar] [CrossRef] [PubMed]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. Mint, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012, 40, D857–D861. [Google Scholar] [CrossRef] [PubMed]
- Cowley, M.J.; Pinese, M.; Kassahn, K.S.; Waddell, N.; Pearson, J.V.; Grimmond, S.M.; Biankin, A.V.; Hautaniemi, S.; Wu, J. Pina v2.0: Mining interactome modules. Nucleic Acids Res. 2012, 40, D862–D865. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Salwinski, L.; Miller, C.S.; Smith, A.J.; Pettit, F.K.; Bowie, J.U.; Eisenberg, D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004, 32, D449–D451. [Google Scholar] [CrossRef] [PubMed]
- Launay, G.; Salza, R.; Multedo, D.; Thierry-Mieg, N.; Ricard-Blum, S. Matrixdb, the extracellular matrix interaction database: Updated content, a new navigator and expanded functionalities. Nucleic Acids Res. 2015, 43, D321–D327. [Google Scholar] [CrossRef] [PubMed]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. Innatedb: Systems biology of innate immunity and beyond—Recent updates and continuing curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Nanduri, B. Hpidb—A unified resource for host-pathogen interactions. BMC Bioinform. 2010, 11 (Suppl. 6), S16. [Google Scholar] [CrossRef] [PubMed]
- Mosca, R.; Ceol, A.; Aloy, P. Interactome3d: Adding structural details to protein networks. Nat. Methods 2013, 10, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, A.; Flammini, A.; Maritan, A.; Vespignani, A. Global protein function prediction from protein-protein interaction networks. Nat. Biotechnol. 2003, 21, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Hishigaki, H.; Nakai, K.; Ono, T.; Tanigami, A.; Takagi, T. Assessment of prediction accuracy of protein function from protein—Protein interaction data. Yeast 2001, 18, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.C.; Hogan, J.D.; Cusick, M.E.; Davis, B.P.; Fancher, A.M.; Hodges, P.E.; Kondu, P.; Lengieza, C.; Lew-Smith, J.E.; Lingner, C.; et al. The yeast proteome database (YPD) and caenorhabditis elegans proteome database (wormpd): Comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000, 28, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabasi, A.L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed]
- Mosca, R.; Tenorio-Laranga, J.; Olivella, R.; Alcalde, V.; Ceol, A.; Soler-Lopez, M.; Aloy, P. Dsysmap: Exploring the edgetic role of disease mutations. Nat. Methods 2015, 12, 167–168. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tibiche, C.; Fu, C.; Kaneko, T.; Moran, M.F.; Schiller, M.R.; Li, S.S.; Wang, E. The human phosphotyrosine signaling network: Evolution and hotspots of hijacking in cancer. Genome Res. 2012, 22, 1222–1230. [Google Scholar] [CrossRef] [PubMed]
- Juan, D.; Pazos, F.; Valencia, A. High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proc. Natl. Acad. Sci. USA 2008, 105, 934–939. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Mason, S.P.; Barabasi, A.L.; Oltvai, Z.N. Lethality and centrality in protein networks. Nature 2001, 411, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the s. Cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.W.; Kern, A.D. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 2005, 22, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Wuchty, S. Interaction and domain networks of yeast. Proteomics 2002, 2, 1715–1723. [Google Scholar] [CrossRef]
- Han, J.D.; Bertin, N.; Hao, T.; Goldberg, D.S.; Berriz, G.F.; Zhang, L.V.; Dupuy, D.; Walhout, A.J.; Cusick, M.E.; Roth, F.P.; et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004, 430, 88–93. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006, 2, e88. [Google Scholar] [CrossRef] [PubMed]
- Zotenko, E.; Mestre, J.; O’Leary, D.P.; Przytycka, T.M. Why do hubs in the yeast protein interaction network tend to be essential: Reexamining the connection between the network topology and essentiality. PLoS Comput. Biol. 2008, 4, e1000140. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, M.; Zhang, Y.; Wang, D.; Li, R.; Guimera, R.; Gao, J.T.; Zhang, M.Q. Modulerole: A tool for modulization, role determination and visualization in protein-protein interaction networks. PLoS ONE 2014, 9, e94608. [Google Scholar] [CrossRef] [PubMed]
- Del Sol, A.; Arauzo-Bravo, M.J.; Amoros, D.; Nussinov, R. Modular architecture of protein structures and allosteric communications: Potential implications for signaling proteins and regulatory linkages. Genome Biol. 2007, 8, R92. [Google Scholar] [CrossRef] [PubMed]
- Ma’ayan, A. Insights into the organization of biochemical regulatory networks using graph theory analyses. J. Biol. Chem. 2009, 284, 5451–5455. [Google Scholar] [CrossRef] [PubMed]
- Crespo, I.; Krishna, A.; Le Bechec, A.; del Sol, A. Predicting missing expression values in gene regulatory networks using a discrete logic modeling optimization guided by network stable states. Nucleic Acids Res. 2013, 41, e8. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Crespo, I.; Androsova, G.; del Sol, A. Discrete logic modelling optimization to contextualize prior knowledge networks using prunet. PLoS ONE 2015, 10, e0127216. [Google Scholar] [CrossRef] [PubMed]
- Osterhout, R.E.; Figueroa, I.A.; Keasling, J.D.; Arkin, A.P. Global analysis of host response to induction of a latent bacteriophage. BMC Microbiol. 2007, 7, 82. [Google Scholar] [CrossRef] [PubMed]

| Database | URL | Number of Interactions * | Number of Species | Data Source * | Ref. |
|---|---|---|---|---|---|
| BIND | http://bind.ca | 58,266 | 10 | E | [79] |
| BioGRID | http://thebiogrid.org | 1,055,196 | 30 | E | [80] |
| HGPD | http://www.hgpd.jp/ | 34,624 | human | E | [81] |
| HPRD | http://www.hprd.org/ | 38,167 | human | E | [17] |
| PIPs | http://www.compbio.dundee.ac.uk/www-pips | 79,441 | human | C | [16] |
| I2D | http://ophid.utoronto.ca/ | 687,072 (E) 619,398 (C) | 6 | E; C | [82] |
| HitPredict | http://hintdb.hgc.jp/htp/ | 398,696 | 105 | E | [83] |
| IID | http://ophid.utoronto.ca/iid | 1,566,043 | 6 | E; C | [84] |
| IntAct | http://www.ebi.ac.uk/intact | 586,731 | 275 | E | [85] |
| iRefWeb | http://wodaklab.org/iRefWeb/ | 263,479 | 1,448 | E | [86] |
| MINT | http://mint.bio.uniroma2.it/mint/ | 235,635 | ~30 | E | [87] |
| PAIR | http://www.cls.zju.edu.cn/pair/ | 5990 (E) 145,494 (C) | A. thaliana | C, E | [36] |
| PINA | http://cbg.garvan.unsw.edu.au/pina/ | 365,930 | 6 | E | [88] |
| AtPID | http://atpid.biosino.org/ | 28,062 | A. thaliana | C | [33] |
| STRING | http://string-db.org | 919,186,040 | 2,031 | C | [89] |
| DIP | http://dip.doembi.ucla.edu | 81,067 | 808 | E | [90] |
| MatrixDB | http://matrixdb.ibcp.fr | 9,851 | Human | E | [91] |
| InnateDB | http://www.innatedb.com | 227,297 (E) 248,205 (C) | Human Bovine Mouse | C, E | [92] |
| HPIDB | http://www.agbase.msstate.edu/hpi/main.html | 45,482 | 70 host and 598 pathogens | C, E | [93] |
| Interactome 3D | http://interactome3d.irbbarcelona.org/ | 12,000 | 8 | C | [94] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, T.; Peng, W.; Wang, Q.; Wang, B.; Sun, J. Reconstruction and Application of Protein–Protein Interaction Network. Int. J. Mol. Sci. 2016, 17, 907. https://doi.org/10.3390/ijms17060907
Hao T, Peng W, Wang Q, Wang B, Sun J. Reconstruction and Application of Protein–Protein Interaction Network. International Journal of Molecular Sciences. 2016; 17(6):907. https://doi.org/10.3390/ijms17060907
Chicago/Turabian StyleHao, Tong, Wei Peng, Qian Wang, Bin Wang, and Jinsheng Sun. 2016. "Reconstruction and Application of Protein–Protein Interaction Network" International Journal of Molecular Sciences 17, no. 6: 907. https://doi.org/10.3390/ijms17060907
APA StyleHao, T., Peng, W., Wang, Q., Wang, B., & Sun, J. (2016). Reconstruction and Application of Protein–Protein Interaction Network. International Journal of Molecular Sciences, 17(6), 907. https://doi.org/10.3390/ijms17060907