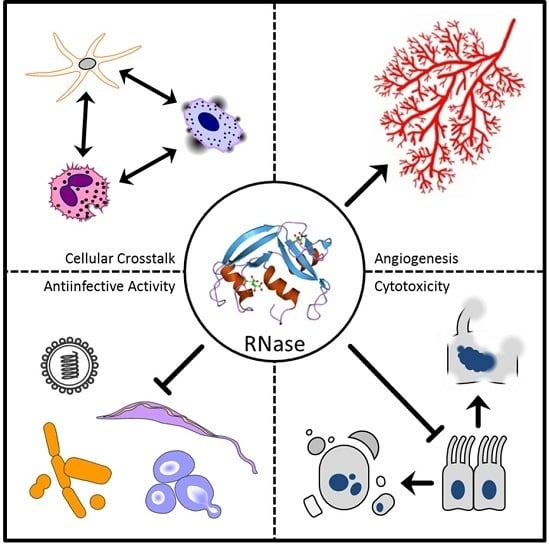
The Ribonuclease A Superfamily in Humans: Canonical RNases as the Buttress of Innate Immunity
Abstract
:1. Introduction
2. Ribonuclease (RNase) 1
3. RNase 2
4. RNase 3
5. RNase 4
6. RNase 5
7. RNase 6
8. RNase 7
9. RNase 8
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| RNase A | Ribonuclease A |
| RI | Ribonuclease inhibitor |
| RNA | Ribonucleic acid |
| DNA | Deoxyribonucleic acid |
| HIV | Human immunodeficiency virus |
| RSV | Respiratory syncytial virus |
| TNFα | Tumour necrosis factor α |
| IL | Interleukin |
| EDN | Eosinophil-derived neurotoxin |
| LPS | Lipopolysaccharide |
| TLR | Toll-like receptor |
| NOD | Nucleotide-binding oligomerization domain |
| Th2 | Type 2 helper T |
| ECP | Eosinophil cationic protein |
| E. coli | Escherichia coli |
| EGFR | Epidermal growth factor receptor |
| NFκB | Nuclear factor κ-light-chain enhancer of activated B cells |
| STAT | Signal transducer and activator of transcription |
| MAPK | Mitogen-activated protein kinase |
| IFNγ | Interferon γ |
| VRE | Vancomycin-resistant enterococci |
| OprI | Outer membrane protein I |
References
- Beintema, J.J.; Kleinedam, R.G. The ribonuclease A superfamily: General discussion. Cell. Mol. Life Sci. 1998, 54, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, S. The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010, 584, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Luhtala, N.; Parker, R. T2 Family ribonucleases: Ancient enzymes with diverse roles. Trends Biochem. Sci. 2010, 35, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Pulido, D.; Valle, J.; Nogués, M.V.; Andreu, D.; Boix, E. Ribonucleases as a host-defence family: Evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem. J. 2013, 456, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Batra, J.K. Antimicrobial activity of human eosinophil granule proteins: Involvement in host defence against pathogens. Crit. Rev. Microbiol. 2012, 38, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F. Eosinophil-derived neurotoxin (EDN/RNase 2) and the mouse eosinophil-associated RNases (mEars): Expanding roles in promoting host defense. Int. J. Mol. Sci. 2015, 16, 15442–15455. [Google Scholar] [CrossRef] [PubMed]
- Boix, E.; Nogués, M.V. Mammalian antimicrobial proteins and peptides: Overview on the RNase A superfamily members involved in innate host defence. Mol. Biosyst. 2007, 3, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Dubois, J.Y.; Ursing, B.M.; Kolkman, J.A.; Beintema, J.J. Molecular evolution of mammalian ribonucleases 1. Mol. Phylogenet. Evol. 2003, 27, 453–463. [Google Scholar] [CrossRef]
- Rosenberg, H.F. RNase A ribonucleases and host defense: An evolving story. J. Leukoc. Biol. 2008, 83, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Premzl, M. Comparative genomic analysis of eutherian ribonuclease A genes. Mol. Genet. Genom. 2014, 289, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Dyer, K.D.; Rosenberg, H.F. The RNase a superfamily: Generation of diversity and innate host defense. Mol. Divers. 2006, 10, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Beintema, J.J.; Zhang, J. The ribonuclease A superfamily of mammals and birds: Identifying new members and tracing evolutionary histories. Genomics 2005, 85, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Buonanno, P.; di Maro, A.; Ponticelli, S.; de Falco, S.; Quarto, N.; Cubellis, M.V.; D’Alessio, G. Ribonucleases and angiogenins from fish. J. Biol. Chem. 2006, 281, 27454–27460. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Zhang, J. Zebrafish ribonucleases are bactericidal: Implications for the origin of the vertebrate RNase A superfamily. Mol. Biol. Evol. 2007, 24, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Rutkoski, T.J.; Raines, R.T. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr. Pharm. Biotechnol. 2008, 9, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, S.; Naddeo, M.; Russo, A.; D’Alessio, G. Degradation of double-stranded RNA by human pancreatic ribonuclease: Crucial role of noncatalytic basic amino acid residues. Biochemistry 2003, 42, 10182–10190. [Google Scholar] [CrossRef] [PubMed]
- Zernecke, A.; Preissner, K.T. Extracellular ribonucleic acids (RNA) enter the stage in cardiovascular disease. Circ. Res. 2016, 118, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Lee-Huang, S.; Huang, P.L.; Sun, Y.; Huang, P.L.; Kung, H.F.; Blithe, D.L.; Chen, H.C. Lysozyme and RNases as anti-HIV components in β-core preparations of human chorionic gonadotropin. Proc. Natl. Acad. Sci. USA 1999, 96, 2678–2681. [Google Scholar] [CrossRef] [PubMed]
- Rugeles, M.T.; Trubey, C.M.; Bedoya, V.I.; Pinto, L.A.; Oppenheim, J.J.; Rybak, S.M.; Shearer, G.M. Ribonuclease is partly responsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. Aids 2003, 17, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Bedoya, V.I.; Boasso, A.; Hardy, A.W.; Rybak, S.; Shearer, G.M.; Rugeles, M.T. Ribonucleases in HIV type 1 inhibition: Effect of recombinant RNases on infection of primary T cells and immune activation-induced RNase gene and protein expression. AIDS Res. Hum. Retrovir. 2006, 22, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Rosenberg, H.F.; Rybak, S.M.; Newton, D.L.; Wang, Z.Y.; Fu, Q.; Tchernev, V.T.; Wang, M.; Schweitzer, B.; et al. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J. Immunol. 2004, 173, 6134–6142. [Google Scholar] [CrossRef] [PubMed]
- Domachowske, J.B.; Dyer, K.D.; Bonville, C.A.; Rosenberg, H.F. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis. 1998, 177, 1458–1464. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D.; Domachowske, J.B. Respiratory viruses and eosinophils: Exploring the connections. Antivir. Res. 2009, 83, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gansler, J.; Preissner, K.T.; Fischer, S. Influence of proinflammatory stimuli on the expression of vascular ribonuclease 1 in endothelial cells. FASEB J. 2014, 28, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D.; Foster, P.S. Eosinophils: Changing perspectives in health and disease. Nat. Rev. Immunol. 2013, 13, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Lacy, P.; Moqbel, R. Signaling and degranulation. In Eosinophils in Health and Disease; Lee, J.J., Rosenberg, H.F., Eds.; Elsevier Press: Waltham, MA, USA, 2013; pp. 206–219. [Google Scholar]
- Abu-Ghazaleh, R.I.; Dunnette, S.L.; Loegering, D.A.; Checkel, J.L.; Kita, H.; Thomas, L.L.; Gleich, G.J. Eosinophil granule proteins in peripheral blood granulocytes. J. Leukoc. Biol. 1992, 52, 611–618. [Google Scholar] [PubMed]
- Hosoki, K.; Nakamura, A.; Nagao, M.; Hiraguchi, Y.; Tokuda, R.; Wada, H.; Nobori, T.; Fujisawa, T. Differential activation of eosinophils by “probiotic” Bifidiobacterium bifidum and “pathogenic” Clostridium difficle. Int. Arch. Allergy Immunol. 2010, 152, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Hosoki, K.; Nakamura, A.; Kainuma, K.; Sugimoto, M.; Nagao, M.; Hiraguchi, Y.; Tanida, H.; Tokuda, R.; Wada, H.; Nobori, T.; et al. Differential activation of eosinophils by bacteria associated with asthma. Int. Arch. Allergy Immunol. 2013, 161, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Su, S.; Zhang, P.; Kurosaka, K.; Caspi, R. Eosinophil-derived neurotoxin acts as an alarmin to activate the TLR2–MyD88 signal pathway in dendritic cells and enhances Th2 immune responses. J. Exp. Med. 2008, 205, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Rosenberg, H.F.; Chen, Q.; Dyer, K.D.; Kurosaka, K.; Oppenheim, J.J. Eosinophil-derived neurotoxin (EDN), an antimicrobial protein with chemotactic activities for dendritic cells. Blood 2003, 102, 3396–3403. [Google Scholar] [CrossRef] [PubMed]
- Domachowske, J.B.; Dyer, K.D.; Adams, A.G.; Leto, T.L.; Rosenberg, H.F. Eosinophil cationic protein/RNase 3 is another RNase A-family ribonuclease with direct antiviral activity. Nucleic Acids Res. 1998, 26, 3358–3363. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Moussaoui, M.; Nogués, M.V.; Torrent, M.; Boix, E. Towards the rational design of antimicrobial proteins single point mutations can switch on bactericidal and agglutinating activities on the RNase A superfamily lineage. FEBS J. 2013, 280, 5841–5852. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Torrent, M.; Andreu, D.; Nogués, M.V.; Boix, E. Two human host defense ribonucleases against mycobacteria, the eosinophil cationic protein (RNase 3) and RNase 7. Antimicrob. Agents Chemother. 2013, 57, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Venge, P.; Byström, J.; Carlson, M.; Hâkansson, L.; Karawacjzyk, M.; Peterson, C.; Sevéus, L.; Trulson, A. Eosinophil cationic protein (ECP): Molecular and biological properties and the use of ECP as a marker of eosinophil activation in disease. Clin. Exp. Allergy 1999, 29, 1172–1186. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, S.J.; Gleich, G.J.; Logering, D.A.; Richardson, B.A.; Butterworth, A.E. Comparative toxicity of purified human eosinophil granule cationic proteins for schistosomula of Schistosoma mansoni. Am. J. Trop. Med. Hyg. 1985, 34, 735–745. [Google Scholar] [PubMed]
- Eriksson, J.; Reimert, C.M.; Kabatereine, N.B.; Kazibwe, F.; Ireri, E.; Kadzo, H.; Eltahir, H.B.; Mohamed, A.O.; Vennervald, B.J.; Venge, P. The 434(G>C) polymorphism within the coding sequence of eosinophil cationic protein (ECP) correlates with the natural course of Schistosoma mansoni infection. Int. J. Parasitol. 2007, 37, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Adu, B.; Dodoo, D.; Adukpo, S.; Gyan, B.A.; Hedley, P.L.; Goka, B.; Adjei, G.O.; Larsen, S.O.; Christiansen, M.; Theisen, M. Polymorphisms in the RNASE3 gene are associated with susceptibility to cerebral malaria in ghanaian children. PLoS ONE 2011, 6, e29465. [Google Scholar] [CrossRef] [PubMed]
- Hamann, K.; Gleich, G.; Checkel, J.; Loegering, D.; McCall, J.; Barker, R. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J. Immunol. 1990, 144, 3166–3173. [Google Scholar] [PubMed]
- Hamann, K.; Barker, R.; Loegering, D.; Gleich, G. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis. J. Parasitol. 1987, 73, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Kitazoe, M.; Tada, H.; de Llorens, R.; Salomon, D.S.; Ueda, M. Growth inhibition of mammalian cells by eosinophil cationic protein. Eur. J. Biochem. 2002, 269, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Carreras, E.; Boix, E.; Rosenberg, H.F.; Cuchillo, C.M.; Nogués, M.V. Both aromatic and cationic residues contribute to the membranelytic and bactericidal activity of eosinophil cationic protein. Biochemistry 2003, 42, 6636–6644. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D. Human ribonuclease 4 (RNase 4): Coding sequence, chromosomal localization and identification of two distinct transcripts in human somatic tissues. Nucleic Acids Res. 1995, 23, 4290–4295. [Google Scholar] [CrossRef] [PubMed]
- Egesten, A.; Dyer, K.D.; Batten, D.; Domachowske, J.B.; Rosenberg, H.F. Ribonucleases and host defense: Identification, localization and gene expression in adherent monocytes in vitro. Biochim. Biophys. Acta 1997, 1358, 255–260. [Google Scholar] [CrossRef]
- Futami, J.; Tsushima, Y.; Murato, Y.; Tada, H.; Sasaki, J.; Seno, M.; Yamada, H. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997, 16, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Abtin, A.; Eckhart, L.; Mildner, M.; Ghannadan, M.; Harder, J.; Schröder, J.M.; Tschachler, E. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J. Investig. Dermatol. 2009, 129, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Koutroubakis, I.E.; Xidakis, C.; Karmiris, K.; Sfiridaki, A.; Kandidaki, E.; Kouroumalis, E.A. Serum angiogenin in inflammatory bowel disease. Dig. Dis. Sci. 2004, 49, 1758–1762. [Google Scholar] [CrossRef] [PubMed]
- Nittoh, T.; Hirakata, M.; Mue, S.; Ohuchi, K. Identification of cDNA encoding rat eosinophil cationic protein/eosinophil-associated ribonuclease. Biochim. Biophys. Acta 1997, 1351, 42–46. [Google Scholar] [CrossRef]
- Spear, G.T.; Kendrick, S.R.; Chen, H.Y.; Thomas, T.T.; Bahk, M.; Balderas, R.; Ghosh, S.; Weinberg, A.; Landay, A.L. Multiplex immunoassay of lower genital tract mucosal fluid from women attending an urban STD clinic shows broadly increased IL1β and lactoferrin. PLoS ONE 2011, 6, e19560. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Stappenbeck, T.S.; Hong, C.V.; Gordon, J.I. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003, 4, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Avdeeva, S.V.; Chernukha, M.U.; Shaginyan, I.A.; Tarantul, V.Z.; Naroditsky, B.S. Human angiogenin lacks specific antimicrobial activity. Curr. Microbiol. 2006, 53, 477–478. [Google Scholar] [CrossRef] [PubMed]
- Kulka, M.; Fukuishi, N.; Metcalfe, D.D. Human mast cells synthesize and release angiogenin, a member of the ribonuclease A (RNase A) superfamily. J. Leukoc. Biol. 2009, 86, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Shcheglovitova, O.; Maksyanina, E.; Ionova, I.; Rustam’yan, Y.L.; Komolova, G. Cow milk angiogenin induces cytokine production in human blood leukocytes. Bull. Exp. Biol. Med. 2003, 135, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Tschesche, H.; Kopp, C.; Hörl, W.; Hempelmann, U. Inhibition of degranulation of polymorphonuclear leukocytes by angiogenin and its tryptic fragment. J. Biol. Chem. 1994, 269, 30274–30280. [Google Scholar] [PubMed]
- Schmaldienst, S.; Oberpichler, A.; Tschesche, H.; Hörl, W.H. Angiogenin: A novel inhibitor of neutrophil lactoferrin release during extracorporeal circulation. Kidney Blood Press. Res. 2000, 26, 107–112. [Google Scholar] [CrossRef]
- Becknell, B.; Eichler, T.E.; Beceiro, S.; Li, B.; Easterling, R.S.; Carpenter, A.R.; James, C.L.; McHugh, K.M.; Hains, D.S.; Partida-Sanchez, S.; et al. Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int. 2015, 87, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Arranz-Trullén, J.; Prats-Ejarque, G.; Velázquez, D.; Torrent, M.; Moussaoui, M.; Boix, E. Insights into the antimicrobial mechanism of action of human RNase6: Structural determinants for bacterial cell agglutination and membrane permeation. Int. J. Mol. Sci. 2016, 17, 552. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.D.; Schwaderer, A.L.; Eichler, T.; Wang, H.; Kline, J.; Justice, S.S.; Cohen, D.M.; Hains, D.S. An endogenous ribonuclease inhibitor regulates the antimicrobial activity of ribonuclease 7 in the human urinary tract. Kidney Int. 2014, 85, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Laudien, M.; Dressel, S.; Harder, J.; Gläser, R. Differential expression pattern of antimicrobial peptides in nasal mucosa and secretion. Rhinology 2011, 49, 107–111. [Google Scholar] [PubMed]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Götz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Investig. Dermatol. 2010, 131, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Simanski, M.; Rademacher, F.; Schröder, L.; Schumacher, H.M.; Gläser, R.; Harder, J. IL-17A and IFN-γ synergistically induce RNase 7 expression via STAT3 in primary keratinocytes. PLoS ONE 2013, 8, e59531. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Wu, S.J.; Chang, T.W.; Wang, C.F.; Suen, C.S.; Hwang, M.J.; et al. Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein. J. Biol. Chem. 2010, 285, 8985–8994. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Niwata, Y.; Ohgi, K.; Ogawa, M.; Irie, M. Distribution of two urinary ribonuclease-like enzymes in human organs and body fluids. J. Biochem. 1986, 99, 17–25. [Google Scholar] [PubMed]
- Iwama, M.; Kunihiro, M.; Ohgi, K.; Irie, M. Purification and properties of human urine ribonucleases. J. Biochem. 1981, 89, 1005–1016. [Google Scholar] [PubMed]
- De Prisco, R.; Sorrentino, S.; Leone, E.; Libonati, M. A ribonuclease from human seminal plasma active on double-stranded RNA. Biochim. Biophys. Acta 1984, 788, 356–363. [Google Scholar] [CrossRef]
- Yasuda, T.; Nadano, D.; Takeshita, H.; Kishi, K. Two distinct secretory ribonucleases from human cerebrum: Purification, characterization and relationships to other ribonucleases. Biochem. J. 1993, 296, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Mizuta, K.; Awazu, S.; Yasuda, T.; Kishi, K. Purification and characterization of three ribonucleases from human kidney: Comparison with urine ribonucleases. Arch. Biochem. Biophys. 1990, 281, 144–151. [Google Scholar] [CrossRef]
- Landré, J.B.; Hewett, P.W.; Olivot, J.M.; Friedl, P.; Ko, Y.; Sachinidis, A.; Moenner, M. Human endothelial cells selectively express large amounts of pancreatic-type ribonuclease (RNase 1). J. Cell. Biochem. 2002, 86, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Nishio, M.; Dadkhahi, S.; Gansler, J.; Saffarzadeh, M.; Shibamiyama, A.; Kral, N.; Baal, N.; Koyama, T.; Deindl, E.; et al. Expression and localisation of vascular ribonucleases in endothelial cells. Thromb. Haemost. 2011, 105, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.A. Biological function of pancreatic ribonuclease. Nature 1969, 221, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Kannemeier, C.; Shibamiya, A.; Nakazawa, F.; Trusheim, H.; Ruppert, C.; Markart, P.; Song, Y.; Tzima, E.; Kennerknecht, E.; Niepmann, M.; et al. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc. Natl. Acad. Sci. USA 2007, 104, 6388–6393. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Gerriets, T.; Wessels, C.; Walberer, M.; Kostin, S.; Stolz, E.; Zheleva, K.; Hocke, A.; Hippenstiel, S.; Preissner, K.T. Extracellular RNA mediates endothelial-cell permeability via vascular endothelial growth factor. Blood 2007, 110, 2457–2465. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Grantzow, T.; Pagel, J.I.; Tschernatsch, M.; Sperandio, M.; Preissner, K.T.; Deindl, E. Extracellular RNA promotes leukocyte recruitment in the vascular system by mobilising proinflammatory cytokines. Thromb. Haemost. 2012, 108, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Fuentes, H.A.; Lopez, M.L.; McCurdy, S.; Fischer, S.; Meiler, S.; Baumer, Y.; Galuska, S.P.; Preissner, K.T.; Boisvert, W.A. Regulation of monocyte/macrophage polarisation by extracellular RNA. Thromb. Haemost. 2015, 113, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Fuentes, H.A.; Niemann, B.; Grieshaber, P.; Wollbrueck, M.; Gehron, J.; Preissner, K.T.; Böning, A. RNase1 as a potential mediator of remote ischaemic preconditioning for cardioprotection. Eur. J. Cardio-Thorac. Surg. 2015, 48, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.H. Remarks on Hodgkin’s disease: A pathogenic agent in the glands, and its application in diagnosis. Br. Med. J. 1933, 1, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Durack, D.T.; Sumi, S.M.; Klebanoff, S.J. Neurotoxicity of human eosinophils. Proc. Natl. Acad. Sci. USA 1979, 76, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.M.; Bonville, C.A.; Rosenberg, H.F.; Domachowske, J.B. Respiratory syncytical virus-induced chemokine expression in the lower airways: Eosinophil recruitment and degranulation. Am. J. Respir. Crit. Care Med. 1999, 159, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, E.A.; Ochkur, S.I.; Lee, N.A.; Lee, J.J. Eosinophils and asthma. Curr. Allergy Asthma Rep. 2007, 7, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Kvarnhammar, A.M.; Cardell, L.O. Pattern recognition receptors in human eosinophils. Immunology 2012, 136, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Szklarek, D.; Barton, A.; Ganz, T.; Hamann, K.J.; Gleich, G.J. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol. 1989, 142, 4428–4434. [Google Scholar] [PubMed]
- Rosenberg, H.F. Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem. 1995, 270, 7876–7881. [Google Scholar] [CrossRef] [PubMed]
- Molina, H.A.; Kierszenbaum, F.; Hamann, K.J.; Gleich, G.J. Toxic effects produced or mediated by human eosinophil granule components on Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 1988, 38, 327–334. [Google Scholar] [PubMed]
- Boix, E.; Salazar, V.A.; Torrent, M.; Pulido, D.; Nogués, M.V.; Moussaoui, M. Structural determinants of the eosinophil cationic protein antimicrobial activity. Biol. Chem. 2012, 393, 801–815. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Pulido, D.; Nogués, M.V.; Boix, E. Exploring new biological functions of amyloids: Bacteria cell agglutination mediated by host protein aggregation. PLoS Pathog. 2012, 8, e1003005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisa, D.; Alonso, R.; Rábano, A.; Rodal, I.; Carrasco, L. Different brain regions are infected with fungi in Alzheimer’s disease. Sci. Rep. 2015, 5, 15015. [Google Scholar] [CrossRef] [PubMed]
- Young, J.D.E.; Peterson, C.G.B.; Venge, P.; Cohn, Z.A. Mechanism of membrane damage mediated by human eosinophil cationic protein. Nature 1986, 321, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Cuyás, E.; Carreras, E.; Navarro, S.; López, O.; de la Maza, A.; Nogués, M.V.; Reshetnyak, Y.K.; Boix, E. Topography studies on the membrane interaction mechanism of the eosinophil cationic protein. Biochemistry 2007, 46, 720–733. [Google Scholar]
- Torrent, M.; Navarro, S.; Moussaoui, M.; Nogués, M.V.; Boix, E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 2008, 47, 3544–3555. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.; Aleu, J.; Jiménez, M.; Boix, E.; Cuchillo, C.; Nogués, M.V. The cytotoxicity of eosinophil cationic protein/ribonuclease 3 on eukaryotic cell lines takes place through its aggregation on the cell membrane. Cell. Mol. Life Sci. 2008, 65, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.C.; Lo, C.W.; Fan, T.; Chang, M.D.T.; Shu, C.W.; Chang, C.H.; Chung, C.T.; Fang, S.L.; Chao, C.C.; Tsai, J.J.; et al. TNF-α Mediates eosinophil cationic protein-induced apoptosis in BEAS-2 B cells. BMC Cell Biol. 2010, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.C.; Chang, H.T.; Chen, I. A heparan sulfate-facilitated and raft-dependent macropinocytosis of eosinophil cationic protein. Traffic 2007, 8, 1778–1795. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.; Johannessen, K.M.; Smolenski, G.; Callaghan, M.; Broadhurst, M.K.; Kim, K.; Wheeler, T.T. Characterisation of the anti-microbial activity of bovine milk ribonuclease4 and ribonuclease5 (angiogenin). Int. Dairy J. 2010, 20, 400–407. [Google Scholar] [CrossRef]
- Murata, M.; Wakabayashi, H.; Yamauchi, K.; Abe, F. Identification of milk proteins enhancing the antimicrobial activity of lactoferrin and lactoferricin. J. Dairy Sci. 2013, 96, 4891–4898. [Google Scholar] [CrossRef] [PubMed]
- Fett, J.W.; Strydom, D.J.; Lobb, R.R.; Alderman, E.M.; Bethune, J.L.; Riordan, J.F.; Vallee, B.L. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 1985, 24, 5480–5486. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Vallee, B.L. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bond generated by regional mutagenesis. Biochemistry 1989, 28, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.S.; Vallee, B.L. Characterization of ribonucleolytic activity of angiogenin towards tRNA. Biochem. Biophys. Res. Commun. 1989, 161, 121–126. [Google Scholar] [CrossRef]
- Sheng, J.; Xu, Z. Three decades of research on angiogenin: A review and perspective. Acta Biochim. Biophys. Sin. 2016, 48, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, T.; Sun, Y.; Kishimoto, K.; Olson, K.A.; Liu, S.; Hirukawa, S.; Hu, G.F. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 2005, 65, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Lee, I.; Kang, T.C.; Jeong, G.B.; Chang, S.I. Angiogenin is involved in morphological changes and angiogenesis in the ovary. Biochem. Biophys. Res. Commun. 1999, 257, 182–186. [Google Scholar] [CrossRef] [PubMed]
- King, T.V.; Vallee, B.L. Neovascularisation of the meniscus with angiogenin. An experimental study in rabbits. J. Bone Jt. Surg. 1991, 73, 587–590. [Google Scholar]
- Walker, C.R.; Hautefort, I.; Dalton, J.E.; Overweg, K.; Egan, C.E.; Bongaerts, R.J.; Newton, D.J.; Cruickshank, S.M.; Andrew, E.M.; Carding, S.R. Intestinal intraepithelial lymphocyte-enterocyte crosstalk regulates production of bactericidal angiogenin 4 by Paneth cells upon microbial challenge. PLoS ONE 2013, 8, e84553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forman, R.A.; deSchoolmeester, M.L.; Hurst, R.J.; Wright, S.H.; Pemberton, A.D.; Else, K.J. The goblet cell is the cellular source of the anti-microbial angiogenin 4 in the large intestine post Trichuris muris infection. PLoS ONE 2012, 7, e42248. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.; Dyer, K.D. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): Increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res. 1996, 24, 3507–3513. [Google Scholar] [CrossRef] [PubMed]
- Christensen-Quick, A.; Lafferty, M.; Sun, L.; Marchionni, L.; DeVico, A.; Garzino-Demo, A. Human Th17 cells lack HIV-inhibitory RNases and are highly permissive to productive HIV infection. J. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Schröder, J. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002, 277, 46779–46784. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial RNases of human skin. J. Investig. Dermatol. 2009, 129, 2091–2093. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Schröder, J.M. Psoriatic scales: A promising source for the isolation of human skin-derived antimicrobial proteins. J. Leukoc. Biol. 2005, 77, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Firat, Y.H.; Simanski, M.; Rademacher, F.; Schröder, L.; Brasch, J.; Harder, J. Infection of keratinocytes with Trichophytum rubrum induces epidermal growth factor-dependent RNase 7 and human β-defensin-3 expression. PLoS ONE 2014, 9, e93941. [Google Scholar] [CrossRef] [PubMed]
- Otri, A.M.; Mohammed, I.; Abedin, A.; Cao, Z.; Hopkinson, A.; Panjwani, N.; Dua, H.S. Antimicrobial peptides expression by ocular surface cells in response to Acanthamoeba castellanii: An in vitro study. Br. J. Ophthalmol. 2010, 94, 1523–1527. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, I.; Yeung, A.; Abedin, A.; Hopkinson, A.; Dua, H.S. Signalling pathways involved in ribonuclease-7 expression. Cell. Mol. Life Sci. 2010, 68, 1941–1952. [Google Scholar] [CrossRef] [PubMed]
- Burgey, C.; Kern, W.V.; Römer, W.; Sakinc, T.; Rieg, S. The innate defense antimicrobial peptides hBD3 and RNase7 are induced in human umbilical vein endothelial cells by classical inflammatory cytokines but not Th17 cytokines. Microbes Infect. 2015, 17, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Köten, B.; Simanski, M.; Gláser, R.; Podschun, R.; Schröder, J.M.; Harder, J. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS ONE 2009, 4, e6424. [Google Scholar] [CrossRef] [PubMed]
- Fritz, P.; Beck-Jendroschek, V.; Brasch, J. Inhibition of dermatophytes by the antimicrobial peptides human β-defensin-2, ribonuclease 7 and psoriasin. Med. Mycol. 2012, 50, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Badia, M.; Moussaoui, M.; Sanchez, D.; Nogués, M.V.; Boix, E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010, 277, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.C.; Moser, J.M.; Dyer, K.D.; Percopo, C.M.; Rosenberg, H.F. Genetic diversity of human RNase 8. BMC Genom. 2012, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, B.; Podschun, R.; Sahly, H.; Schubert, S.; Schröder, J.M.; Harder, J. Identification of RNase 8 as a novel human antimicrobial protein. Antimicrob. Agents Chemother. 2006, 50, 3194–3196. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Paliou, M.; Hohmann, E.L.; Calderwood, S.B.; Wing, E.J. Listeriosis during pregnancy: A case series and review of 222 cases. Medicine 2002, 81, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Haigh, B.J.; Griffin, F.J.; Wheeler, T.T. The mammalian secreted RNases: Mechanisms of action in host defence. Innate Immun. 2013, 19, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Koczera, P.; Simons, N.; Zechendorf, E.; Hoeger, J.; Marx, G.; Schuerholz, T. The human host defense ribonucleases 1, 3 and 7 are elevated in patients with sepsis after major surgery—A pilot study. Int. J. Mol. Sci. 2016, 17, 294. [Google Scholar] [CrossRef] [PubMed]
- Schuerholz, T.; Brandenburg, K.; Marx, G. Antimicrobial peptides and their potential application in inflammation and sepsis. Crit. Care 2012, 16, 207. [Google Scholar] [CrossRef] [PubMed]



| Ribonuclease | Proposed Impact on Host Defence | Reference(s) |
|---|---|---|
| RNase 1 | Degradation of vascular polyRNA | [18,19] |
| Anti-HIV-1 activity | [20,21,22] | |
| Induces maturation and activation of dendritic cells | [23] | |
| RNase 2/EDN | Antiviral activity against HIV-1 and RSV-B | [20,21,22,24,25] |
| Secretion by eosinophil granulocytes and monocyte-derived macrophages | [23,26,27,28,29,30,31] | |
| TLR2 binding and Th2 polarization | [32] | |
| Chemokine and cytokine induction for activation and maturation of dendritic cells | [23,33] | |
| RNase 3/ECP | Antiviral activity against RSV-B | [34] |
| Antibacterial activity against mycobacteria and Gram+ and Gram− bacteria | [35,36] | |
| Induces degranulation of mast cells | [37] | |
| Anthelmintic activity against Schistosoma mansoni, Brugia pahangi and Trichinella spiralis | [38,39,40,41,42,43] | |
| Cytotoxic activity against mammalian cells | [43,44] | |
| RNase 4 | Expression in host defence-associated tissues | [45,46,47,48] |
| Coexpression with lactoferrin, lactoferricin and RNase 5 Enhances antimicrobial activity of lactoferrin and lactoferricin | ||
| RNase 5/Angiogenin | Increased serum levels during acute-phase response | [49,50,51] |
| Antiviral activity against HIV-1 | [22] | |
| Activity against Candida | [48,52,53] | |
| Activity against Streptococcus (controversial data) | ||
| Synthesis and secretion by mast cells | [54] | |
| Proinflammatory stimulation of leukocytes | [55] | |
| Inhibition of degranulation of neutrophil granulocytes | [56,57] | |
| RNase 6 | Infection-induced secretion in urinary tract | [58,59] |
| Antibacterial activity against Gram+ and Gram− bacteria | ||
| RNase 7 | Synthesis upon microbial, inflammatory and physicochemical challenge in epithelial tissues | [59,60,61,62] |
| Antibacterial activity against mycobacteria and Gram+ and Gram− bacteria | [36,59,63] | |
| RNase 8 | Antibacterial and antifungal activity against Gram+ and Gram− bacteria and Candida | [64] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koczera, P.; Martin, L.; Marx, G.; Schuerholz, T. The Ribonuclease A Superfamily in Humans: Canonical RNases as the Buttress of Innate Immunity. Int. J. Mol. Sci. 2016, 17, 1278. https://doi.org/10.3390/ijms17081278
Koczera P, Martin L, Marx G, Schuerholz T. The Ribonuclease A Superfamily in Humans: Canonical RNases as the Buttress of Innate Immunity. International Journal of Molecular Sciences. 2016; 17(8):1278. https://doi.org/10.3390/ijms17081278
Chicago/Turabian StyleKoczera, Patrick, Lukas Martin, Gernot Marx, and Tobias Schuerholz. 2016. "The Ribonuclease A Superfamily in Humans: Canonical RNases as the Buttress of Innate Immunity" International Journal of Molecular Sciences 17, no. 8: 1278. https://doi.org/10.3390/ijms17081278
APA StyleKoczera, P., Martin, L., Marx, G., & Schuerholz, T. (2016). The Ribonuclease A Superfamily in Humans: Canonical RNases as the Buttress of Innate Immunity. International Journal of Molecular Sciences, 17(8), 1278. https://doi.org/10.3390/ijms17081278