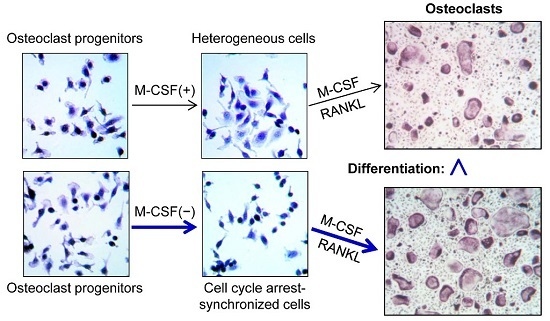
Synchronized Cell Cycle Arrest Promotes Osteoclast Differentiation
Abstract
:1. Introduction
2. Results and Discussion
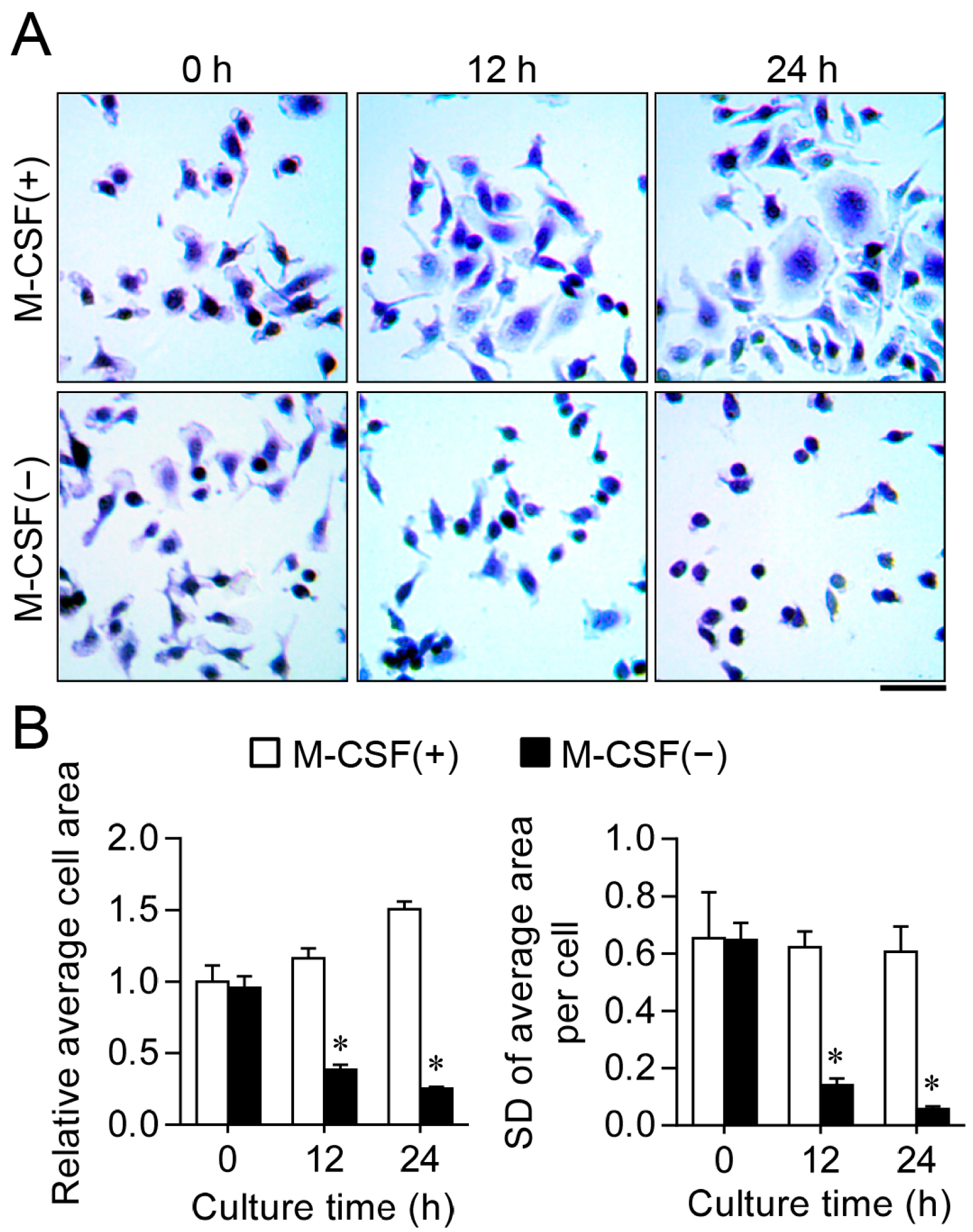
2.1. M-CSF Deprivation Induces G0–G1 Cell Cycle Arrest
2.2. Cell Synchronization Promotes Osteoclast Formation
3. Materials and Methods
3.1. Induction of Synchronized Cell Cycle Arrest and Osteoclast Differentiation
3.2. Analysis of Cell Area
3.3. Assay of Cell Proliferation and Cell Cycle Analysis
3.4. Immunoblot Analysis
3.5. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Myster, D.L.; Duronio, R.J. To differentiate or not to differentiate? Curr. Biol. 2000, 10, R302–R304. [Google Scholar] [PubMed]
- Scott, R.E.; Florine, D.L.; Wille, J.J., Jr.; Yun, K. Coupling of growth arrest and differentiation at a distinct state in the G1 phase of the cell cycle: Gd. Proc. Natl. Acad. Sci. USA 1982, 79, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Vaessin, H. Pan-neural prospero terminates cell proliferation during drosophila neurogenesis. Genes Dev. 2000, 14, 147–151. [Google Scholar] [PubMed]
- Rodier, A.; Marchal-Victorion, S.; Rochard, P.; Casas, F.; Cassar-Malek, I.; Rouault, J.P.; Magaud, J.P.; Mason, D.Y.; Wrutniak, C.; Cabello, G. BTG1: A triiodothyronine target involved in the myogenic influence of the hormone. Exp. Cell Res. 1999, 249, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Halevy, O.; Novitch, B.G.; Spicer, D.B.; Skapek, S.X.; Rhee, J.; Hannon, G.J.; Beach, D.; Lassar, A.B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by myoD. Science 1995, 267, 1018–1021. [Google Scholar] [CrossRef] [PubMed]
- De la Serna, I.L.; Roy, K.; Carlson, K.A.; Imbalzano, A.N. Myod can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 2001, 276, 41486–41491. [Google Scholar] [CrossRef] [PubMed]
- Li, V.C.; Kirschner, M.W. Molecular ties between the cell cycle and differentiation in embryonic stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 9503–9508. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.Y.; Yoshida, H.; Sarosi, I.; Tan, H.L.; Timms, E.; Capparelli, C.; Morony, S.; Oliveira-dos-Santos, A.J.; Van, G.; Itie, A.; et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999, 397, 315–323. [Google Scholar] [PubMed]
- Yoshida, H.; Hayashi, S.; Kunisada, T.; Ogawa, M.; Nishikawa, S.; Okamura, H.; Sudo, T.; Shultz, L.D.; Nishikawa, S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 1990, 345, 442–444. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.C.; Takada, Y.; Shishodia, S.; Aggarwal, B.B. Evidence that receptor activator of nuclear factor (NF)-kappaB ligand can suppress cell proliferation and induce apoptosis through activation of a NF-kappaB-independent and TRAF6-dependent mechanism. J. Biol. Chem. 2004, 279, 6065–6076. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, T.; Katagiri, M.; Yamamoto, A.; Hoshi, K.; Takato, T.; Nakamura, K.; Tanaka, S.; Okayama, H.; Kawaguchi, H. Osteoclast differentiation by RANKL requires NF-kappaB-mediated downregulation of cyclin-dependent kinase 6 (cdk6). J. Bone Miner. Res. 2004, 19, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, T.; Muto, A.; Udagawa, N.; Arai, A.; Yamashita, T.; Hosoya, A.; Ninomiya, T.; Nakamura, H.; Yamamoto, Y.; Kinugawa, S.; et al. Identification of cell cycle-arrested quiescent osteoclast precursors in vivo. J. Cell Biol. 2009, 184, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Dormond, O.; Lejeune, F.J.; Ruegg, C. Modulation of cdk2, cyclin D1, p16INK4a, p21WAF and p27Kip1 expression in endothelial cells by TNF/IFN gamma. Anticancer Res. 2002, 22, 3159–3163. [Google Scholar] [PubMed]
- Nowakowski, R.S.; Lewin, S.B.; Miller, M.W. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J. Neurocytol. 1989, 18, 311–318. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.P.; Osmani, A.H.; Wu, L.P.; Spotts, J.L.; Osmani, S.A. Mitotic histone H3 phosphorylation by the nima kinase in aspergillus nidulans. Cell 2000, 102, 293–302. [Google Scholar] [CrossRef]
- Lee, K.; Chung, Y.H.; Ahn, H.; Kim, H.; Rho, J.; Jeong, D. Selective regulation of mapk signaling mediates RANKL-dependent osteoclast differentiation. Int. J. Biol. Sci. 2016, 12, 235–245. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, M.; Kim, J.-M.; Lee, K.; Park, S.-Y.; Lim, H.-S.; Kim, T.; Jeong, D. Synchronized Cell Cycle Arrest Promotes Osteoclast Differentiation. Int. J. Mol. Sci. 2016, 17, 1292. https://doi.org/10.3390/ijms17081292
Kwon M, Kim J-M, Lee K, Park S-Y, Lim H-S, Kim T, Jeong D. Synchronized Cell Cycle Arrest Promotes Osteoclast Differentiation. International Journal of Molecular Sciences. 2016; 17(8):1292. https://doi.org/10.3390/ijms17081292
Chicago/Turabian StyleKwon, Minsuk, Jin-Man Kim, Kyunghee Lee, So-Young Park, Hyun-Sook Lim, Taesoo Kim, and Daewon Jeong. 2016. "Synchronized Cell Cycle Arrest Promotes Osteoclast Differentiation" International Journal of Molecular Sciences 17, no. 8: 1292. https://doi.org/10.3390/ijms17081292
APA StyleKwon, M., Kim, J.-M., Lee, K., Park, S.-Y., Lim, H.-S., Kim, T., & Jeong, D. (2016). Synchronized Cell Cycle Arrest Promotes Osteoclast Differentiation. International Journal of Molecular Sciences, 17(8), 1292. https://doi.org/10.3390/ijms17081292