MicroRNAs: Potential Biomarkers and Therapeutic Targets for Alveolar Bone Loss in Periodontal Disease
Abstract
:1. Introduction
2. Biogenesis of MicroRNAs (miRNAs)
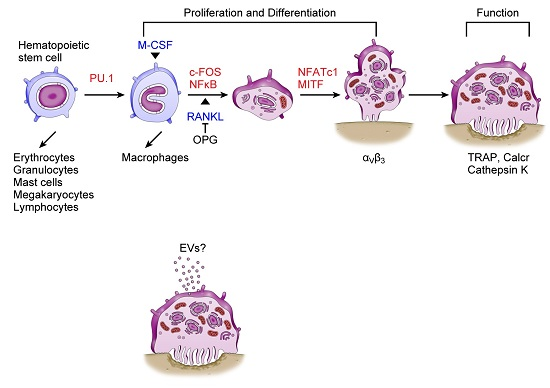
3. Osteoclasts and miRNAs
4. Extracellular miRNAs
5. Materials and Methods
5.1. Bone Marrow Macrophage Culture and Fluorescence Staining of Actin and Nuclei
5.2. Human Peripheral Blood CD14+ Cell Culture and Quantitative RT-PCR Analysis
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Kagiya, T.; Nakamura, S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor α and RANKL during osteoclast differentiation. J. Periodontal Res. 2013, 48, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Kagiya, T. MicroRNAs and osteolytic bone metastasis: The roles of microRNAs in tumor-induced osteoclast differentiation. J. Clin. Med. 2015, 4, 1741–1752. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Takayanagi, H. Osteoclasts and the immune system. J. Bone Miner Metab. 2009, 27, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Kagiya, T. Roles of microRNAs in osteoclast differentiation and function. In Osteoclasts: Cell Biology, Functions and Related Disease; Reeves, C., Ed.; Nova Science Publishers: New York, NY, USA, 2015; pp. 1–18. [Google Scholar]
- Perri, R.; Nares, S.; Zhang, S.; Barros, S.P.; Offenbacher, S. MicroRNA modulation in obesity and periodontitis. J. Dent. Res. 2012, 91, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Na, H.S.; Jeong, S.Y.; Jeong, S.H.; Park, H.R.; Chung, J. Comparison of inflammatory microRNA expression in healthy and periodontitis tissues. Biocell 2011, 35, 43–49. [Google Scholar] [PubMed]
- Xie, Y.F.; Shu, R.; Jiang, S.Y.; Liu, D.L.; Zhang, X.L. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int. J. Oral Sci. 2011, 3, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin-Wasmer, C.; Guarnieri, P.; Celenti, R.; Demmer, R.T.; Kebschull, M.; Papapanou, P.N. MicroRNAs and their target genes in gingival tissues. J. Dent. Res. 2012, 91, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Ogata, Y.; Matsui, S.; Kato, A.; Zhou, L.; Nakayama, Y.; Takai, H. MicroRNA expression in inflamed and noninflamed gingival tissues from Japanese patients. J. Oral Sci. 2014, 56, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.U.; Miyazaki, H.; Ochiya, T. The roles of microRNAs in breast cancer. Cancers 2015, 7, 598–616. [Google Scholar] [CrossRef] [PubMed]
- Lui, P.Y.; Jin, D.Y.; Stevenson, N.J. MicroRNA: Master controllers of intracellular signaling pathways. Cell. Mol. Life Sci. 2015, 72, 3531–3542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X. Lipopolysaccharide-regulated production of bone sialoprotein and interleukin-8 in human periodontal ligament fibroblasts: The role of toll-like receptors 2 and 4 and the MAPK pathway. J. Periodontal Res. 2015, 50, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 11, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Sugatani, T.; Vacher, J.; Hruska, K.A. A microRNA expression signature of osteoclastogenesis. Blood 2011, 117, 3648–3657. [Google Scholar] [CrossRef] [PubMed]
- Osta, B.; Benedetti, G.; Miossec, P. Classical and paradoxical effects of TNF-α on bone homeostasis. Front. Immunol. 2014, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Kapinas, K.; Kessler, C.B.; Delany, A.M. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical Wnt signaling. J. Cell. Biochem. 2009, 108, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hassan, M.Q.; Jafferji, M.; Aqeilan, R.I.; Garzon, R.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009, 284, 15676–15684. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. MicroRNAs in NF-κB signaling. J. Mol. Cell. Biol. 2011, 3, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Pitari, M.R.; Amodio, N.; di Martino, M.T.; Conforti, F.; Leone, E.; Botta, C.; Paolino, F.M.; del Giudice, T.; Iuliano, E.; et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: Implications for the treatment of multiple myeloma-related bone disease. J. Cell. Physiol. 2013, 228, 1506–1515. [Google Scholar] [CrossRef] [PubMed]
- Franceschetti, T.; Kessler, C.B.; Lee, S.K.; Delany, A.M. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J. Biol. Chem. 2013, 288, 33347–33360. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Weinberg, R.A. miR-31: A crucial overseer of tumor metastasis and other emerging roles. Cell Cycle 2010, 9, 2124–2129. [Google Scholar] [CrossRef] [PubMed]
- Itzstein, C.; Coxon, F.P.; Rogers, M.J. The regulation of osteoclast function and bone resorption by small GTPases. Small GTPases 2011, 2, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, F.; Murakami, Y.; Saito, T.; Miyasaka, N.; Kohsaka, H. miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Res. Ther. 2013, 15, R102. [Google Scholar] [CrossRef] [PubMed]
- Krzeszinski, J.Y.; Wei, W.; Huynh, H.; Jin, Z.; Wang, X.; Chang, T.C.; Xie, X.J.; He, L.; Mangala, L.S.; Lopez-Berestein, G.; et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 2014, 512, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Nakamachi, Y.; Ohnuma, K.; Uto, K.; Noguchi, Y.; Saegusa, J.; Kawano, S. MicroRNA-124 inhibits the progression of adjuvant-induced arthritis in rats. Ann. Rheum. Dis. 2016, 75, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, H.J.; Park, C.K.; Kim, Y.G.; Lee, H.J.; Kim, J.Y.; Kim, H.H. MicroRNA-124 regulates osteoclast differentiation. Bone 2013, 56, 383–389. [Google Scholar] [CrossRef] [PubMed]
- De la Rica, L.; Garcia-Gomez, A.; Comet, N.R.; Rodriguez-Ubreva, J.; Ciudad, L.; Vento-Tormo, R.; Company, C.; Alvarez-Errico, D.; Garcia, M.; Gomez-Vaquero, C.; et al. NF-κB-direct activation of microRNAs with repressive effects on monocyte-specific genes is critical for osteoclast differentiation. Genome Biol. 2015, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.J.; Liao, L.; Yang, L.; Li, Y.; Jiang, T.J. miR-125a TNF receptor-associated factor 6 to inhibit osteoclastogenesis. Exp. Cell Res. 2014, 321, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Nakasa, T.; Shibuya, H.; Nagata, Y.; Niimoto, T.; Ochi, M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011, 63, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Takahashi, N.; Miyauchi, S.; Yamazaki, K. Porphyromonas gingivalis lipopolysaccharide I nduces miR-146a without altering the production of inflammatory cytokines. Biochem. Biophys. Res. Commun. 2012, 420, 918–925. [Google Scholar] [PubMed]
- Bala, S.; Csak, T.; Momen-Heravi, F.; Lippai, D.; Kodys, K.; Catalano, D.; Satishchandran, A.; Ambros, V.; Szabo, G. Biodistribution and function of extracellular miRNA-155 in mice. Sci. Rep. 2015, 5, 10721. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, A.R.; Manjunath, S.G.; Swati, P.P.; Shikha, C.; Sujatha, P.B. Gingival crevicular fluid levels of leukotriene B4 in periodontal health and disease. J. Periodontol. 2007, 78, 2325–2230. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Filgueiras, L.R.; Wang, S.; Serezani, A.P.; Peters-Golden, M.; Jancar, S.; Serezani, C.H. Leukotriene B4 enhances the generation of proinflammatory microRNAs to promote MyD88-dependent macrophage activation. J. Immunol. 2014, 192, 2349–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, M.; Barad, O.; Agami, R.; Geiger, B.; Hornstein, E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc. Natl. Acad. Sci. USA 2010, 107, 15804–15809. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, F.; Izu, Y.; Hayata, T.; Hemmi, H.; Nakashima, K.; Nakamura, T.; Kato, S.; Miyasaka, N.; Ezura, Y.; Noda, M. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J. Cell. Biochem. 2010, 109, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Bluml, S.; Bonelli, M.; Niederreiter, B.; Puchner, A.; Mayr, G.; Hayer, S.; Koenders, M.I.; van den Berg, W.B.; Smolen, J.; Redlich, K. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis Rheum. 2011, 63, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Sugatani, T.; Hruska, K.A. MicroRNA-223 is a key factor in osteoclast differentiation. J. Cell. Biochem. 2007, 101, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Sugatani, T.; Hruska, K.A. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J. Biol. Chem. 2009, 284, 4667–4678. [Google Scholar] [CrossRef] [PubMed]
- M’Baya-Moutoula, E.; Louvet, L.; Metzinger-Le Meuth, V.; Massy, Z.A.; Metzinger, L. High inorganic phosphate concentration inhibits osteoclastogenesis by modulating miR-223. Biochim. Biophys. Acta 2015, 1852, 2202–2212. [Google Scholar] [CrossRef] [PubMed]
- Sugatani, T.; Hruska, K.A. Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J. Cell. Biochem. 2013, 114, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Ell, B.; Mercatali, L.; Ibrahim, T.; Campbell, N.; Schwarzenbach, H.; Pantel, K.; Amadori, D.; Kang, Y. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell 2013, 24, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Chen, C.; He, H.B.; Hu, R.; Zhou, H.D.; Xie, H.; Zhu, W.; Dai, R.C.; Wu, X.P.; Liao, E.Y.; Luo, X.H. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J. Bone Miner Res. 2013, 28, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Lee, S.U.; Kim, E.H.; Park, S.J.; Choi, I.; Kim, T.D.; Kim, S.H. Osteoporotic bone of miR-150-deficient mice: Possibly due to low serum OPG-mediated osteoclast activation. Bone Rep. 2015, 3, 5–10. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, H.; Chen, J.; Xia, B.; Jin, Y.; Wei, W.; Shen, J.; Huang, Y. Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett. 2012, 586, 3255–3262. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Crescitelli, R.; Lasser, C.; Szabo, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzas, E.I.; Lotvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida-Aoki, N.; Ochiya, T. Interactions between cancer cells and normal cells via miRNAs in extracellular vesicles. Cell. Mol. Life Sci. 2015, 72, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Raposo, G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and Intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Kagiya, T.; Taira, M. A new application for microarrays: Analysis of global microRNA expression profiles in the extracellular microvesicles of human macrophage-like cells. In Microarrays: Principles, Applications and Technologies; Rogers, J.V., Ed.; Nova Science Publishers: New York, NY, USA, 2014; pp. 69–80. [Google Scholar]
- Kagiya, T.; Taira, M. Expression of microRNAs in the extracellular microvesicles of murine osteoclasts. J. Oral Tissue Eng. 2013, 10, 142–150. [Google Scholar]
- Ortega, F.J.; Moreno, M.; Mercader, J.M.; Moreno-Navarrete, J.M.; Fuentes-Batllevell, N.; Sabater, M.; Ricart, W.; Fernandez-Real, J.M. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin. Epigenet. 2015, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Yu, M.L.; Yu, G.; Bian, J.J.; Deng, X.M.; Wan, X.J.; Zhu, K.M. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010, 394, 184–188. [Google Scholar] [CrossRef] [PubMed]





| miRNA | Function(s) | Target(s) | Reference(s) | Gingiva with Periodontitis | Reference(s) |
|---|---|---|---|---|---|
| miR-21 | P | Pdcd4, FasL | [17,44] | UP | [8] |
| miR-29b | N | C-FOS, MMP-2 | [22] | UP | [8] |
| miR-29a/b/c | P | Calcr, Cd93, Cdc42, Gpr85, Nfia, Srgap2 | [23] | UP | [8] |
| miR-31 | P | RhoA | [26] | DOWN | [10] |
| miR-34a | N | Tgif2 | [27] | UP/DOWN | [8,11] |
| miR-124 | N | NFATc1 | [28,29] | Not reported | |
| miR-125a | P/N | TRAF6, TNFIP3 | [30,31] | UP | [8] |
| miR-141 | N | Mitf, Calcr | [45] | DOWN | [10] |
| miR-146a | N | TRAF6 | [32] | UP | [9] |
| miR-148a | P | MAFB | [46] | UP | [10] |
| miR-150 | N | Opg | [47] | UP | [11] |
| miR-155 | N | Mitf, Socs1, Pu.1 | [1,38,48] | UP/DOWN | [9,10] |
| miR-223 | P/N | NFI-A | [1,41,42,43] | UP | [10,11] |
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kagiya, T. MicroRNAs: Potential Biomarkers and Therapeutic Targets for Alveolar Bone Loss in Periodontal Disease. Int. J. Mol. Sci. 2016, 17, 1317. https://doi.org/10.3390/ijms17081317
Kagiya T. MicroRNAs: Potential Biomarkers and Therapeutic Targets for Alveolar Bone Loss in Periodontal Disease. International Journal of Molecular Sciences. 2016; 17(8):1317. https://doi.org/10.3390/ijms17081317
Chicago/Turabian StyleKagiya, Tadayoshi. 2016. "MicroRNAs: Potential Biomarkers and Therapeutic Targets for Alveolar Bone Loss in Periodontal Disease" International Journal of Molecular Sciences 17, no. 8: 1317. https://doi.org/10.3390/ijms17081317
APA StyleKagiya, T. (2016). MicroRNAs: Potential Biomarkers and Therapeutic Targets for Alveolar Bone Loss in Periodontal Disease. International Journal of Molecular Sciences, 17(8), 1317. https://doi.org/10.3390/ijms17081317