Cellular Metabolomics Revealed the Cytoprotection of Amentoflavone, a Natural Compound, in Lipopolysaccharide-Induced Injury of Human Umbilical Vein Endothelial Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Assay of NO, Malondialdehyde (MDA), and Superoxide Dismutase (SOD) Activity
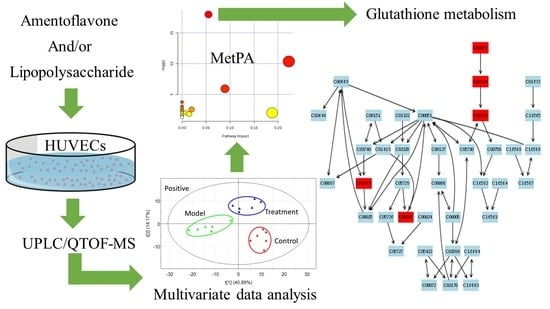
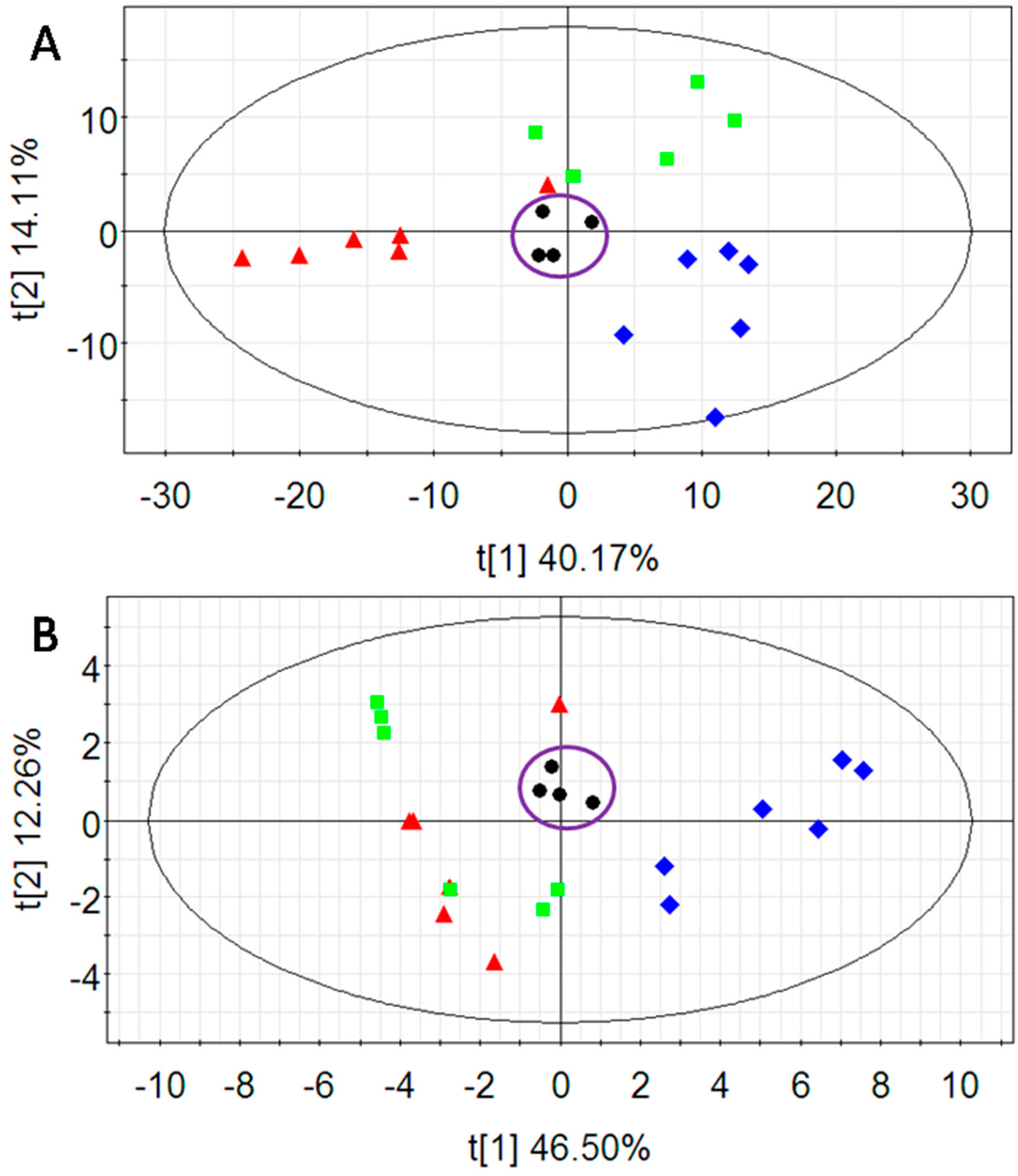
2.2. Multivariate Data Analysis
2.3. Tentative Identification of the Biomarkers and Pathway Analysis
2.4. Effects of Amentoflavone on the Metabolic Pathway
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Preparation of Amentoflavone Solutions
3.3. Cell Culture
3.4. Cell Metabolite Extraction
3.5. Biochemical Assays of SOD Activity, MDA Level and NO Level
3.6. UPLC/QTOF-MS Analysis of Intracellular Metabolites
3.7. Multivariate Data Analysis and Data Processing
3.8. Biomarker Identification and Metabolic Pathway Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LPS | lipopolysaccharide |
| HUVECs | human umbilical vein endothelial cells |
| NO | nitric oxide |
| NOS-2 | type-2 isoform of nitric oxide synthase |
| RNOS | reactive nitrogen oxide species |
| GSH | glutathione |
| GS-NO | S-nitrosoglutothione |
| MDA | malondialdehyde |
| SOD | superoxide dismutase |
| PCA | principal component analysis |
| PLS-DA | projections to latent structures discriminate analysis |
| QC | quality control |
| UV | unit variance |
| VIP | variable importance in the projection |
| R2X | the cumulative fraction of sum of squares of X explained by components |
| R2Y | the cumulative sum of squares of all the y-variables explained by the extracted components |
| Q2Y | the fraction of the total variation of Y that can be predicted by the extracted components |
| ANOVA | analysis of variance |
| UPLC/QTOF-MS | Ultra-performance liquid chromatography/quadrupole time of flight-mass spectrometry |
References
- Li, M.; Li, H.; Jiang, P.; Liu, X.; Xu, D.; Wang, F. Investigating the pathological processes of rhegmatogenous retinal detachment and proliferative vitreoretinopathy with metabolomics analysis. Mol. BioSyst. 2014, 10, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Irie, M.; Fujimura, Y.; Yamato, M.; Miura, D.; Wariishi, H. Integrated MALDI-MS imaging and LC-MS techniques for visualizing spatiotemporal metabolomic dynamics in a rat stroke model. Metabolomics 2014, 10, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Wang, M.X.; Chowbay, B.; Ching, C.B.; Chen, W.N. Metabolic profiling of HepG2 cells incubated with S(−) and R(+) enantiomers of anti-coagulating drug warfarin. Metabolomics 2011, 7, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, Y.; Meng, X.; Yin, P.; Deng, C.; Chen, J.; Wang, Z.; Xu, G. Effect of a traditional Chinese medicine preparation Xindi soft capsule on rat model of acute blood stasis: A urinary metabonomics study based on liquid chromatography–mass spectrometry. J. Chromatogr. B 2008, 873, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Antti, H.; Ebbels, T.M.; Keun, H.C.; Bollard, M.E.; Beckonert, O.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. Statistical experimental design and partial least squares regression analysis of biofluid metabonomic NMR and clinical chemistry data for screening of adverse drug effects. Chemom. Intell. Lab. Syst. 2004, 73, 139–149. [Google Scholar] [CrossRef]
- Panopoulos, A.D.; Yanes, O.; Ruiz, S.; Kida, Y.S.; Diep, D.; Tautenhahn, R.; Herrerias, A.; Batchelder, E.M.; Plongthongkum, N.; Lutz, M.; et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012, 22, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Halama, A. Metabolomics in cell culture—A strategy to study crucial metabolic pathways in cancer development and the response to treatment. Arch. Biochem. Biophys. 2014, 564, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.-M.; Uppal, K.; Walker, D.I.; Tran, V.; Dury, L.; Strobel, F.H.; Baubichon-Cortay, H.; Pennell, K.D.; Roede, J.R.; Jones, D.P. Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. Methods. Mol. Biol. 2014, 1198, 43–73. [Google Scholar] [PubMed]
- The National Pharmacopoeia Commission of China. Pharmacopoeia of the People’s Republic of China; Chinese Medical Science and Technology Press: Beijing, China, 2010; Volume 1, p. 200. [Google Scholar]
- Shan, M.Q.; Shang, J.; Ding, A.W. Platycladus orientalis leaves: A systemic review on botany, phytochemistry and pharmacology. Am. J. Chin. Med. 2014, 42, 523–542. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.N.; Yao, W.F.; Liu, J.; Shang, J.; Shan, M.Q.; Zhang, L.; Ding, A.W. Protective effect of different solvent extracts from Platycladi Cacumen Carbonisatum on LPS-induced human umbilical vein endothelial cells damage. Zhongguo Zhong Yao Za Zhi 2013, 38, 3933–3938. [Google Scholar] [PubMed]
- Chen, L.; Ding, L.; Yu, A.; Yang, R.; Wang, X.; Li, J.; Jin, H.; Zhang, H. Continuous determination of total flavonoids in Platycladus orientalis (L.) Franco by dynamic microwave-assisted extraction coupled with on-line derivatization and ultraviolet-visible detection. Anal. Chim. Acta 2007, 596, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Paya, M.; Rios, J.; Alcaraz, M. Structure-activity relationships of polymethoxyflavones and other flavonoids as inhibitors of non-enzymic lipid peroxidation. Biochem. Pharmacol. 1990, 40, 793–797. [Google Scholar] [CrossRef]
- Lu, Y.-H.; Liu, Z.-Y.; Wang, Z.-T.; Wei, D.-Z. Quality evaluation of Platycladus orientalis (L.) Franco through simultaneous determination of four bioactive flavonoids by high-performance liquid chromatography. J. Pharm. Biomed. Anal. 2006, 41, 1186–1190. [Google Scholar] [CrossRef] [PubMed]
- Nijveldt, R.J.; van Nood, E.; van Hoorn, D.E.; Boelens, P.G.; van Norren, K.; van Leeuwen, P.A. Flavonoids: A review of probable mechanisms of action and potential applications. Am. J. Clin. Nutr. 2001, 74, 418–425. [Google Scholar] [PubMed]
- Lin, Y.M.; Anderson, H.; Flavin, M.T.; Pai, Y.H.; Mata-Greenwood, E.; Pengsuparp, T.; Pezzuto, J.M.; Schinazi, R.F.; Hughes, S.H.; Chen, F.C. In vitro anti-HIV activity of biflavonoids isolated from Rhus succedanea and Garcinia multiflora. J. Nat. Prod. 1997, 60, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.G.; Yin, M.H.; Oh, H.; Lee, D.H.; Lee, H.S. Vasorelaxation by amentoflavone isolated from Selaginella tamariscina. Planta Med. 2004, 70, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Sellick, C.A.; Knight, D.; Croxford, A.S.; Maqsood, A.R.; Stephens, G.M.; Goodacre, R.; Dickson, A.J. Evaluation of extraction processes for intracellular metabolite profiling of mammalian cells: Matching extraction approaches to cell type and metabolite targets. Metabolomics 2010, 6, 427–438. [Google Scholar] [CrossRef]
- Tennenberg, S.D.; Weller, J.J. Endotoxin-induced, neutrophil-mediated endothelial cytotoxicity is enhanced by T-lymphocytes. J. Surg. Res. 1997, 69, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Biffl, W.L.; Moore, E.E.; Zallen, G.; Johnson, J.L.; Gabriel, J.; Offner, P.J.; Silliman, C.C. Neutrophils are primed for cytotoxicity and resist apoptosis in injured patients at risk for multiple organ failure. Surgery 1999, 126, 198–202. [Google Scholar] [CrossRef]
- Guzik, T.J.; Korbut, R.; Adamek-Guzik, T. Nitric oxide and superoxide in inflammation. J. Physiol. Pharmacol. 2003, 54, 469–487. [Google Scholar] [PubMed]
- Loh, K.P.; Qi, J.; Tan, B.K.H.; Liu, X.H.; Wei, B.G.; Zhu, Y.Z. Leonurine protects middle cerebral artery occluded rats through antioxidant effect and regulation of mitochondrial function. Stroke 2010, 41, 2661–2668. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Hu, W.; Wu, H.; Hu, X. No evident dose-response relationship between cellular ROS level and its cytotoxicity-A paradoxical issue in ROS-based cancer therapy. Sci. Rep. 2014, 4, 5029. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.W. Nitric oxide: A regulator of mast cell activation and mast cell-mediated inflammation. Clin. Exp. Immunol. 2002, 129, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Wink, D.A.; Hanbauer, I.; Grisham, M.B.; Laval, F.; Nims, R.W.; Laval, J.; Cook, J.; Pacelli, R.; Liebmann, J.; Krishna, M.; et al. Chemical biology of nitric oxide: Regulation and protective and toxic mechanisms. Curr. Top. Cell Regul. 1996, 34, 159–187. [Google Scholar] [PubMed]
- Meister, A.; Anderson, M.E. Glutathione. Annu. Rev. Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef] [PubMed]
- Requintina, P.J.; Oxenkrug, G.F. Differential effects of lipopolysaccharide on lipid peroxidation in F344N, SHR rats and BALB/c mice, and protection of melatonin and NAS against its toxicity. Ann. N. Y. Acad. Sci. 2003, 993, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Sahu, B.D.; Rentam, K.K.; Putcha, U.K.; Kuncha, M.; Vegi, G.M.; Sistla, R. Carnosic acid attenuates renal injury in an experimental model of rat cisplatin-induced nephrotoxicity. Food Chem. Toxicol. 2011, 49, 3090–3097. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhao, J.; Ma, L.; Ding, Y.; Su, D. Hepatoprotective effects of total triterpenoids and total flavonoids from Vitis vinifera L against immunological liver injury in mice. Evid. Based Complement. Altern. Med. 2012, 2012, 969386. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Kettaneh-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi-and Megavariate Data Analysis: Part I: Basic Principles and Applications, 2nd ed.; Umetrics Academy: Umeå, Sweden, 2006; p. 97. [Google Scholar]
- Musiek, E.S.; Yin, H.; Milne, G.L.; Morrow, J.D. Recent advances in the biochemistry and clinical relevance of the isoprostane pathway. Lipids 2005, 40, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Kim, J.D.; Lee, W.Y.; Chung, B.C.; Choi, M.H. Quantitative metabolic profiling of 21 endogenous corticosteroids in urine by liquid chromatography–triple quadrupole-mass spectrometry. Anal. Chim. Acta 2009, 632, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; de Marzo, A.M.; van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [PubMed]
- Chaiswing, L.; Oberley, T.D. Extracellular/microenvironmental redox state. Antioxid. Redox Signal. 2010, 13, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Jongkind, J.F.; Verkerk, A.; Baggen, R.G. Glutathione metabolism of human vascular endothelial cells under peroxidative stress. Free Radic. Biol. Med. 1989, 7, 507–512. [Google Scholar] [CrossRef]
- Woo, E.R.; Lee, J.Y.; Cho, I.J.; Kim, S.G.; Kang, K.W. Amentoflavone inhibits the induction of nitric oxide synthase by inhibiting NF-κB activation in macrophages. Pharmacol. Res. 2005, 51, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Cynober, L.A. Metabolic and Therapeutic Aspects of Amino Acids in Clinical Nutrition, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2003; p. 135. [Google Scholar]
- Bender, D.A. Amino Acid Metabolism, 3rd ed.; Wiley-Blackwell: Chichester, UK, 2012; p. 15. [Google Scholar]
- Shanmugam, S.; Sathish, K.T.; Selvam, P. Laboratory Handbook on Biochemistry; PHI Learning Private Limited: Delhi, India, 2010; p. 12. [Google Scholar]
- Lu, Z.H.; Zhang, R.; Diasio, R.B. Purification and characterization of dihydropyrimidine dehydrogenase from human liver. J. Biol. Chem. 1992, 267, 17102–17109. [Google Scholar] [PubMed]
- Yokota, H.; Fernandez-Salguero, P.; Furuya, H.; Lin, K.; McBride, O.W.; Podschun, B.; Schnackerz, K.D.; Gonzalez, F.J. cDNA cloning and chromosome mapping of human dihydropyrimidine dehydrogenase, an enzyme associated with 5-fluorouracil toxicity and congenital thymine uraciluria. J. Biol. Chem. 1994, 269, 23192–23196. [Google Scholar] [PubMed]
- Gemelli, T.; de Andrade, R.B.; Rojas, D.B.; Bonorino, N.F.; Mazzola, P.N.; Tortorelli, L.S.; Funchal, C.; Filho, C.S.; Wannmacher, C.M. Effects of β-alanine administration on selected parameters of oxidative stress and phosphoryltransfer network in cerebral cortex and cerebellum of rats. Mol. Cell. Biochem. 2013, 380, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ramirez, A.; Ortiz-Balderas, E.; Cardozo-Saldana, G.; Diaz-Diaz, E.; El-Hafidi, M. Glycine restores glutathione and protects against oxidative stress in vascular tissue from sucrose-fed rats. Clin. Sci. 2014, 126, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, J.M.; Varani, J.; Johnson, K.J.; Roeser, N.F.; Dame, M.K.; Davis, J.A.; Venkatachalam, M.A. Protection of human umbilical vein endothelial cells by glycine and structurally similar amino acids against calcium and hydrogen peroxide-induced lethal cell injury. Am. J. Pathol. 1992, 140, 457–471. [Google Scholar] [PubMed]
- Wang, Y.; Gao, D.; Chen, Z.; Li, S.; Gao, C.; Cao, D.; Liu, F.; Liu, H.; Jiang, Y. Acridone derivative 8a induces oxidative stress-mediated apoptosis in CCRF-CEM leukemia cells: Application of metabolomics in mechanistic studies of antitumor agents. PLoS ONE 2013, 8, e63572. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. The KEGG database. Novartis Found. Symp. 2002, 247, 91–101. [Google Scholar] [PubMed]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef] [PubMed]




| Group | Concentration/μM | NO/μmol·L−1 | MDA/nmol·mL−1 | SOD/U·mL−1 |
|---|---|---|---|---|
| Vehicle control | – | 21.03 ± 0.86 | 1.71 ± 0.13 | 21.69 ± 2.16 |
| Model | – | 53.66 ± 3.07 * | 2.80 ± 0.26 * | 11.11 ± 1.20 * |
| Treatment | 4.647 | 31.62 ± 3.01 # | 2.13 ± 0.13 # | 18.12 ± 0.50 # |
| 9.294 | 27.33 ± 1.79 # | 1.93 ± 0.09 # | 19.26 ± 0.62 # | |
| 18.587 | 22.13 ± 2.28 # | 1.68 ± 0.17 # | 22.21 ± 1.67 # |
| Methods | Mode | Features | PLS-DA | ||||
|---|---|---|---|---|---|---|---|
| A a | R2X | R2Y | Q2Y | Difference b | |||
| MZmine 2.10 | Positive | 1342 | 3 | 0.375 | 0.936 | 0.579 | 0.357 |
| Negative | 241 | 3 | 0.431 | 0.908 | 0.617 | 0.291 | |
| ANOVA (p < 0.05) c | Positive | 307 | 5 | 0.684 | 0.991 | 0.843 | 0.148 |
| Negative | 31 | 2 | 0.569 | 0.831 | 0.735 | 0.096 | |
| Mode | No. | R.T. (min) a | Mass (m/z) | Adduct Ion | Delta | Formula | Compound | Trend b | Trend c | Cellular Location d |
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | 1 | 0.343 | 113.9899 | M + K | 0.0053 | C2H5NO2 | Glycine | ↓ | ↑ | Mitochondria |
| 2 | 0.738 | 308.1522 | M + NH4 | 0.0043 | C10H18N4O6 | Argininosuccinic acid | ↓ | ↑ | Cytoplasm | |
| 3 | 4.988 | 127.0632 | M + K | 0.0000 | C4H12N2 | Putrescine | ↓ | ↑ | Mitochondria | |
| 4 | 13.744 | 133.0936 | M + H | 0.0035 | C5H12N2O2 | Ornithine | ↓ | ↑ | Mitochondria | |
| 5 | 14.671 | 163.1976 | M + NH4 | 0.0058 | C7H19N3 | Spermidine | ↓ | ↑ | Cytoplasm | |
| Negative | 1 | 0.733 | 128.0383 | M − H | 0.0030 | C5H7NO3 | 5-oxoproline | ↓ | ↑ | Cytoplasm |
| 2 | 14.850 | 113.0293 | M − H | 0.0063 | C4H6N2O2 | Dihydrouracil | ↑ | ↓ | Cytoplasm |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, W.; Li, H.; Liu, Q.; Gao, Y.; Dai, J.; Bao, B.; Zhang, L.; Ding, A. Cellular Metabolomics Revealed the Cytoprotection of Amentoflavone, a Natural Compound, in Lipopolysaccharide-Induced Injury of Human Umbilical Vein Endothelial Cells. Int. J. Mol. Sci. 2016, 17, 1514. https://doi.org/10.3390/ijms17091514
Yao W, Li H, Liu Q, Gao Y, Dai J, Bao B, Zhang L, Ding A. Cellular Metabolomics Revealed the Cytoprotection of Amentoflavone, a Natural Compound, in Lipopolysaccharide-Induced Injury of Human Umbilical Vein Endothelial Cells. International Journal of Molecular Sciences. 2016; 17(9):1514. https://doi.org/10.3390/ijms17091514
Chicago/Turabian StyleYao, Weifeng, Hui Li, Qinan Liu, Ye Gao, Jin Dai, Beihua Bao, Li Zhang, and Anwei Ding. 2016. "Cellular Metabolomics Revealed the Cytoprotection of Amentoflavone, a Natural Compound, in Lipopolysaccharide-Induced Injury of Human Umbilical Vein Endothelial Cells" International Journal of Molecular Sciences 17, no. 9: 1514. https://doi.org/10.3390/ijms17091514
APA StyleYao, W., Li, H., Liu, Q., Gao, Y., Dai, J., Bao, B., Zhang, L., & Ding, A. (2016). Cellular Metabolomics Revealed the Cytoprotection of Amentoflavone, a Natural Compound, in Lipopolysaccharide-Induced Injury of Human Umbilical Vein Endothelial Cells. International Journal of Molecular Sciences, 17(9), 1514. https://doi.org/10.3390/ijms17091514