MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases
Abstract
:1. Introduction
2. Results
2.1. Overexpression of miR-29a Alleviated Fibrosis in Cholestatic Livers
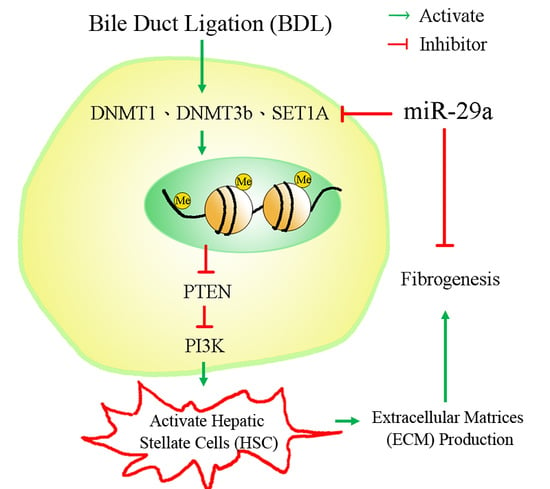
2.2. miR-29a Overexpression Reduced DNA Methyltransferases, Histone Methyltransferase and SET Domain Containing 1A in Cholestatic Mice
2.3. miR-29a Overexpression Increased PTEN and Lowered PI3K Signaling in Cholestatic Livers
2.4. miR-29a Regulated DNA Methylation and Histone Methyltransferases Expressions in Hepatic Stellate Cells
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Construction and Breeding of the miR-29a Transgenic Mouse Colony
4.3. Animal Model and Experimental Protocol
4.4. Primary HSC Isolation and Culture
4.5. RNAi Transfection
4.6. RNA Isolation and Real-Time Quantitative RT-PCR
4.7. Immunohistological Analysis
4.8. Western Blot Analysis
4.9. Genetic Methylation Study
4.10. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, S.C.; Wang, F.S.; Yang, Y.L.; Tiao, M.M.; Chuang, J.H.; Huang, Y.H. Microarray study of pathway analysis expression profile associated with microRNA-29a with regard to murine cholestatic liver injuries. Int. J. Mol. Sci. 2016, 17, 324. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology 2008, 134, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L. Evolving challenges in hepatic fibrosis. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Chuang, J.H.; Yang, Y.L.; Huang, C.C.; Wu, C.L.; Chen, C.L. Cholestasis downregulate hepcidin expression through inhibiting IL-6-induced phosphorylation of signal transducer and activator of transcription 3 signaling. Lab. Investig. 2009, 89, 1128–1139. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Tiao, M.M.; Huang, L.T.; Chuang, J.H.; Kuo, K.C.; Yang, Y.L.; Wang, F.S. Activation of miR-29a in activated hepatic stellate cells modulates its profibrogenic phenotype through inhibition of histone deacetylases 4. PLoS ONE 2015, 10, e0136453. [Google Scholar] [CrossRef] [PubMed]
- Tiao, M.M.; Wang, F.S.; Huang, L.T.; Chuang, J.H.; Kuo, H.C.; Yang, Y.L.; Huang, Y.H. MicroRNA-29a protects against acute liver injury in a mouse model of obstructive jaundice via inhibition of the extrinsic apoptosis pathway. Apoptosis 2014, 19, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Friedman, R.C.; Marquez, R.T.; Keck, K.; Kong, B.; Icardi, M.S.; Brown, K.E.; Burge, C.B.; Schmidt, W.N.; Wang, Y.; et al. Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: miR-29 overexpression reduces hepatitis C viral abundance in culture. J. Infect. Dis. 2011, 203, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Sheen-Chen, S.M.; Lin, C.R.; Chen, K.H.; Yang, C.H.; Lee, C.T.; Huang, H.W.; Huang, C.Y. Epigenetic histone methylation regulates transforming growth factor β-1 expression following bile duct ligation in rats. J. Gastroenterol. 2014, 49, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Perugorria, M.J.; Wilson, C.L.; Zeybel, M.; Walsh, M.; Amin, S.; Robinson, S.; White, S.A.; Burt, A.D.; Oakley, F.; Tsukamoto, H.; et al. Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology 2012, 56, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zheng, L.; Xie, S.; Yin, F.; Huo, X.; Guo, J.; Zhang, X. Regulatory effects and mechanism of adenovirus-mediated PTEN gene on hepatic stellate cells. Dig. Dis. Sci. 2016, 61, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Gubbins, J.; Peng, Z.; Medina, V.; Fei, F.; Asahina, K.; Wang, J.; Kahn, M.; Rountree, C.B.; Stiles, B.L. Activation of hepatic stellate cell in PTEN null liver injury model. Fibrogenes. Tissue Repair 2016, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Bian, E.B.; Huang, C.; Ma, T.T.; Tao, H.; Zhang, H.; Cheng, C.; Lv, X.W.; Li, J. DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell activation and liver fibrogenesis in rats. Toxicol. Appl. Pharmacol. 2012, 264, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Chappell, G.; Kutanzi, K.; Uehara, T.; Tryndyak, V.; Hong, H.H.; Hoenerhoff, M.; Beland, F.A.; Rusyn, I.; Pogribny, I.P. Genetic and epigenetic changes in fibrosis-associated hepatocarcinogenesis in mice. Int. J. Cancer 2014, 134, 2778–2788. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.; Mann, D.A. Epigenetics in liver disease: From biology to therapeutics. Gut 2016. [Google Scholar] [CrossRef] [PubMed]
- Tennakoon, A.H.; Izawa, T.; Wijesundera, K.K.; Murakami, H.; Katou-Ichikawa, C.; Tanaka, M.; Golbar, H.M.; Kuwamura, M.; Yamate, J. Immunohistochemical characterization of glial fibrillary acidic protein (GFAP)-expressing cells in a rat liver cirrhosis model induced by repeated injections of thioacetamide (TAA). Exp. Toxicol. Pathol. 2015, 67, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.S.; Power, B.E.; Molloy, P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta 2007, 1775, 138–162. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Tao, H.; Deng, Z.Y.; Lu, C.; Li, J. Non-coding RNA-mediated epigenetic regulation of liver fibrosis. Metabolism 2015, 64, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Robaina, M.C.; Mazzoccoli, L.; Arruda, V.O.; Reis, F.R.; Apa, A.G.; de Rezende, L.M.; Klumb, C.E. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp. Mol. Pathol. 2015, 98, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Galli, R.; Paone, A.; Fabbri, M.; Zanesi, N.; Calore, F.; Cascione, L.; Acunzo, M.; Stoppacciaro, A.; Tubaro, A.; Lovat, F.; et al. Toll-like receptor 3 (TLR3) activation induces microRNA-dependent reexpression of functional RARbeta and tumor regression. Proc. Natl. Acad. Sci. USA 2013, 110, 9812–9817. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Bakky, M.S.; Hammad, M.A.; Walker, L.A.; Ashfaq, M.K. Tissue factor dependent liver injury causes release of retinoid receptors (RXR-α and RAR-α) as lipid droplets. Biochem. Biophys. Res. Commun. 2011, 410, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, H.; Zhu, N.L.; Wang, J.; Asahina, K.; Machida, K. Morphogens and hepatic stellate cell fate regulation in chronic liver disease. J. Gastroenterol. Hepatol. 2012, 27, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, C.; Lin, Z.; Guo, Y.; Shi, L.; Dong, P.; Lu, Z.; Gao, S.; Liao, Y.; Chen, B.; et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation--a novel mechanism suppressing liver fibrosis. FEBS J. 2014, 281, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Roll, F.J.; Boyles, J.; Bissell, D.M. Hepatic lipocytes: The principal collagen-producing cells of normal rat liver. Proc. Natl. Acad. Sci. USA 1985, 82, 8681–8685. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.H.; Huang, Y.H.; Lin, T.M.; Du, Y.Y.; Tsai, P.C.; Hsieh, C.S.; Chuang, J.H. Selective activation of Toll-like receptor 7 in activated hepatic stellate cells may modulate their profibrogenic phenotype. Biochem. J. 2012, 447, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Kwiecinski, M.; Noetel, A.; Elfimova, N.; Trebicka, J.; Schievenbusch, S.; Strack, I.; Molnar, L.; von Brandenstein, M.; Tox, U.; Nischt, R.; et al. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction. PLoS ONE 2011, 6, e24568. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Schon, H.T.; Bartneck, M.; Borkham-Kamphorst, E.; Nattermann, J.; Lammers, T.; Tacke, F.; Weiskirchen, R. Pharmacological intervention in hepatic stellate cell activation and hepatic fibrosis. Front. Pharmacol. 2016, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Koyama, Y.; Brenner, D.A. New therapies for hepatic fibrosis. Clin. Res. Hepatol. Gastroenterol. 2015, 39, S75–S79. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.; Jung, Y. MicroRNAs in liver fibrosis: Focusing on the interaction with hedgehog signaling. World J. Gastroenterol. 2016, 22, 6652–6662. [Google Scholar] [CrossRef] [PubMed]








© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-L.; Wang, F.-S.; Li, S.-C.; Tiao, M.-M.; Huang, Y.-H. MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases. Int. J. Mol. Sci. 2017, 18, 192. https://doi.org/10.3390/ijms18010192
Yang Y-L, Wang F-S, Li S-C, Tiao M-M, Huang Y-H. MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases. International Journal of Molecular Sciences. 2017; 18(1):192. https://doi.org/10.3390/ijms18010192
Chicago/Turabian StyleYang, Ya-Ling, Feng-Sheng Wang, Sung-Chou Li, Mao-Meng Tiao, and Ying-Hsien Huang. 2017. "MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases" International Journal of Molecular Sciences 18, no. 1: 192. https://doi.org/10.3390/ijms18010192
APA StyleYang, Y.-L., Wang, F.-S., Li, S.-C., Tiao, M.-M., & Huang, Y.-H. (2017). MicroRNA-29a Alleviates Bile Duct Ligation Exacerbation of Hepatic Fibrosis in Mice through Epigenetic Control of Methyltransferases. International Journal of Molecular Sciences, 18(1), 192. https://doi.org/10.3390/ijms18010192