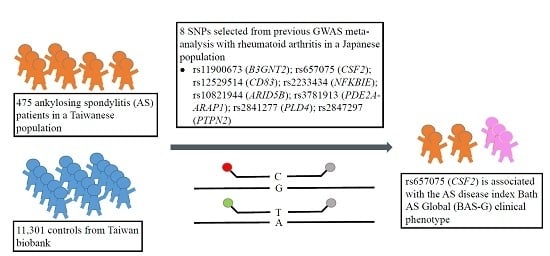
1. Introduction
Ankylosing spondylitis (AS) is a systemic autoimmune disease, which is characterized by inflammation of the lumbar spine and sacroiliac joints, peripheral inflammatory arthropathy, and the absence of rheumatoid factor [
1,
2]. The disease predominantly strikes men between the age of 20 and 40 years, in their peak productive years, leading to significant loss of work productivity and a decreased quality of life [
3]. Family and twin studies indicated that genetic factors contribute to over 90% to the overall AS susceptibility [
4,
5].
Human leukocyte antigen (HLA)-B 27 has been known as the major AS-susceptibility gene for more than 40 years [
6], but
HLA-B27 accounts for only 16% of the genetic variability in AS [
7]. The other
HLA-B allele operative in AS susceptibility is
HLA-B60, which was identified in studies of US and UK patients with AS [
8,
9]. In addition,
HLA-B60 and
HLA-B61 were associated with AS in
HLA-B27-negative patients in Taiwan [
10]. The susceptibility to AS can be enhanced when combining different patterns of
HLA-B60 and
HLA-B27 genotypes in Dutch and Taiwanese populations, implying genetic interaction mechanisms that may contribute to AS risk among two genes [
11,
12]. The
interleukin (IL)-1 and
IL-23R genes were also proven to be important in the pathogenesis of AS [
13,
14]. Genes involved in regulating peripheral tolerance were found have a combined effect on the development of AS, such as the
PD-1 G-536A/
PD-L1 A8923C/
PD-L2 C47103T [
15] or
PTPN22 G-1123C/
CTLA-4 A49G [
16], and the
CTLA-4 +49A/G genotype associated with circulatory C-reactive protein (CRP) level [
17]. Our previous studies reported that genetic polymorphisms of
ORAI1 (rs12313273 and rs7135617) and
STIM1 (rs3750996) were associated with the pathogenesis of
HLA-B27-positive AS patients [
18,
19].
Additionally, many non-major histocompatibility complex (MHC) regions were found to be significantly associated with AS in genome-wide association studies (GWASs) [
20,
21,
22]. Twelve loci were previously confirmed to be associated with AS in Europeans [
20,
21,
23], 2 loci were recently reported in Han Chinese [
22], and an additional 13 new loci were identified in a recent global GWAS, bringing the total AS-associated loci to 43 [
24]. Two studies confirmed the findings of previous AS studies that
ERAP1 and rs10865331 are risk factors for AS susceptibility [
25,
26]. However, some susceptibility loci, such as
EDIL3,
HAPLN1, and
ANO6, discovered in a Han Chinese GWAS were not associated in a Taiwanese AS population [
27].
Rheumatoid arthritis (RA) is a type of autoimmune arthritis, triggered by a faulty immune system resulting in chronic synovitis and the destruction of localized cartilage and bone [
28]. Previous twin and family-base studies indicate that the associations with major histocompatibility complex (MHC) regions explain around 12% of total heritability in the susceptibility of RA [
29]. A recent GWAS meta-analysis indicated that nine new loci were associated with RA in a Japanese population [
30]. Because AS and RA are autoimmune diseases that may share similar genetic factors as well as immune regulatory pathways, we therefore examined if the RA susceptibility polymorphisms from GWAS meta-analysis are also associated with the pathogenesis of AS. In this study, we selected eight single-nucleotide polymorphisms (SNPs) from previous GWAS meta-analysis study. The AS activity index (Bath AS Disease Activity Index, BASDAI), Bath AS Functional Index (BASFI), and Bath AS Global (BAS-G), as well as inflammatory biochemical parameters (immunoglobulin A, IgA, erythrocyte sedimentation rate, ESR, and CRP) were analyzed. We found that the AA genotype of rs657075 (
CSF2) was significantly associated to the clinical phenotype Bath AS Global (BAS-G).
3. Discussion
AS and RA are autoimmune diseases with distinct phenotypes. RA is characterized as a chronic inflammatory joint disease with cartilage and bone damage, whereas spondyloarthropathies in AS are illustrated by entheses and subchondral bone marrow inflammation, particularly abnormal osteoproliferation at involved sites. Familial aggregations of AS and RA have been known for years [
31,
32]. In a 2008 Sweden study, Sundquist and coworkers analyzed 30 years of hospitalizations (1973–2004) for concordant and discordant associations among RA, AS, and systemic lupus erythematosus (SLE). They reported that significant concordant measures with standardized incidence ratios (SIRs) or sibling risks for RA and AS were 5.12 and 17.14, respectively. However, the discordant association between RA and AS was not significant, and AS was only associated with AS when discordance was taken into account [
31]. A further study in 2009 indicated that offspring of parental probands with RA were significantly associated with AS with a SIR of 2.96 [
32]. This laid the foundation for the identification of common genetic components for RA and AS. Recently, some autoimmune disease loci were identified as being shared among multiple autoimmune diseases [
33,
34]. Sirota and coworkers (2009) compared genetic variation profiles of six autoimmune disease and found that RA and AS displayed an autoimmune disease locus cluster that was distinct from the others. For instance, the G allele of rs2076530 in
BTNL2 predisposed patients to RA, AS, and type 1 diabetes, but may play a protective role instead in multiple sclerosis and autoimmune thyroid disease.
To date, more than 43 AS risky loci [
24] and 101 RA risky loci [
35] have been identified. Most of the identified risk loci for autoimmune diseases are related to B-cell or T-cell activation pathways, differentiation, or innate immunity, or involved cytokine signaling or regulating peripheral tolerance.
HLA-B27 has been known for years to be the major AS-susceptibility gene [
6], but fewer than 5% of
HLA-B27 carriers develop AS [
24], suggesting that non-
HLA-B27 alleles are important for AS susceptibility. Indeed, we recently provided evidence that genetic polymorphisms of
ORAI1 (rs12313273 and rs7135617) and
STIM1 (rs3750996) were associated with the pathogenesis of
HLA-B27-positive AS patients [
18,
19]. There is evidence that three of the SNPs (rs4552569, rs17095830, and rs13210693) which correspond to two newly identified AS risky loci (
EDIL3-
HAPLN1 and
ANO6) in Han Chinese [
22] were not replicated in Caucasian populations [
20,
21,
23] and were negatively associated with Taiwanese AS subjects in a previous study of ours [
27], eliciting the question of whether these two newly-identified loci associated with AS in Han Chinese [
22] may have been over-represented. Indeed the association of one SNP, rs13210693 at the 6q21 locus, did not reach a
p value at the genome-wide significance level (
p > 5 × 10
−8) [
22].
Bath ankylosing spondylitis global score (BAS-G) was a valuable quantitative measure which gives a global assessment of the well-being of the person with AS over a given time period as completed by the patient. It was first reported by Jones et al. in 1960 [
36], and endorsed by Assessment of SpondyloArthritis international Society (ASAS) [
37]. It uses two horizontal visual analogue scales (10 cm) to measure the effect of AS on the respondent’s well-being, where none = 0 and severe = 10. The first estimates over the last week, and the second over the last six months. In this study, we firstly reported that the genetic polymorphism rs657075 (
CSF2) was associated with BAS-G.
CSF2 loci were previously reported to be associated with RA and exhibited high levels in joints of RA patients [
30]. In this study, for the first time we showed that
CSF2 loci were not associated with AS development, but may be weakly associated with AS clinical phenotype BAS-G levels in a Taiwanese population.
CSF2, also known as granulocyte-macrophage colony-stimulating factor, functions as a cytokine and is secreted by macrophages, T cells, mast cells, natural killer (NK) cells, endothelial cells, and fibroblasts. It stimulates stem cells to produce granulocytes and is part of the immune/inflammatory cascade. On the other hand, the T helper Th17 cells require
Csf2 to induce autoimmune encephalomyelitis or autoimmune neuroinflammation [
38]. It is known that IL-23 and IL-23R are involved in pathogenic Th17 responses, and
IL-23R is associated with many autoimmune diseases such as psoriasis, AS, and Crohn’s disease [
24]. Perhaps levels of the weakly associated AS clinical phenotype BAS-G seen may have something to do with some of these gene–gene interactions.
According to SNAP website, this SNP has high LD with several genes including
CSF2 and
ACSL6 (
Figure 1). Since the GTEx database includes limited cell types, rs657075 does not show significantly associated eQTL with the
CSF2 gene in the current database. It is associated with expression of
ACSL6 in sigmoid colon cells (
Table 6). On searching HaploReg V4.1, rs657075 is involved in changing chromatin status in primary T cells. Therefore, further functional studies are needed to validate the impact of rs657075 on the expression of
CSF2.
Using a high-density immune-related loci platform (Immunochip), Cortes and coworkers reported that some of the AS risky loci overlapped with those of other immune diseases. Eleven of these AS risky loci were positively associated with ulcerative colitis and 12 loci with Crohn’s disease. However, only two of the AS risky loci were marginally associated with RA; rs11065898 (
SH2B3 or
LNK) and rs2283790 (
UBE2L3) displayed a concordant and discordant mode, respectively. As
SH2B3 encoded an adaptor protein involving T cell signaling, it may be functionally related to the pathogenesis of AS. Interestingly, rs11065898 loci were indeed positively associated with CD4
+ lymphocyte counts [
24].
The modest correlation between RA risk SNPs and AS susceptibility that we observed in this study may due to several reasons.
Firstly, the SNPs were identified from a RA GWAS study conducted in a Japanese cohort. In this study, we assess their contribution to AS susceptibility and disease severity in a Taiwanese cohort. The nonsignificant results regarding susceptibility to Taiwanese AS patients may be attributed to the heterogeneity of the phenotype (diagnostic criteria of the disease). In considering of phenotypic heterogeneity, we have noted different criteria of clinical diagnosis between RA (in the GWAS study from a Japanese population) and AS (in this study based on a Taiwanese population).
Secondly, it is likely that different genetic backgrounds (variation in allele frequencies across different ethnic population admixture) in two populations may play a role in the observed inconsistent results. In particular, different genetic backgrounds between Japanese and Taiwanese populations may have an impact on the genetic influence of rs657075 in the two study populations, consequently, this may result in the inconsistent association that we observed in these two studies.
Thirdly, the prevalences of AS between Taiwanese and Japanese populations are different. The prevalence in Japanese population is lower than in Taiwanese population (0.0065% vs. 0.19%–0.54%) [
39,
40,
41]. The distinct prevalence of AS may imply various genetic factors influencing different underlying regulations in AS across population, which may partially explain the nonsignificant association observed in our study. For example, a previous study reported that three SNPs (rs4552569, rs17095830, and rs13210693) were associated with AS in Han Chinese [
22], but this was not replicated in Caucasian population [
20,
21,
23], nor were these associated with Taiwanese AS subjects [
27].
Fourthly, it has been known that the genetic effect size of SNP may be different across populations. Since some important confounding factors are unmeasured and can not be adjusted in the analyses, this may have a certain degree of influence when determining the risk effect of those SNPs on AS.
Although no correlation between RA susceptible SNPs and AS risk was observed, we have shown that these SNPs are good candidates for AS disease severity, i.e., BASDAI, BASFI, and BAS-G after adjusting for the effects of gender, age, and disease duration. It would still be interesting to consider those previously identified SNPs as good candidates for determining increased risk of AS. As such, further investigation would be merited to validate these SNPs identified from the Japanese AS GWAS study. It will be also important to the authors to further examine the association between these SNPs and increasing AS risk when sample size allows in the future.
The small sample size in this study may have limited the statistical power for detecting sizeable changes in AS risk assessments. Larger cohort studies and replication studies in different populations are required to validate our findings in this study. In summary, although the SNPs (rs657075 on CSF2 and rs11900673 on B3GNT2) are not the risk alleles for the susceptibility of AS, we provide supportive evidence that these SNPs are important generic markers for clinical manifestations of AS patients in a Taiwanese population. Further investigation would be needed to validate the findings in this study.