The Role of the S40 Gene Family in Leaf Senescence
Abstract
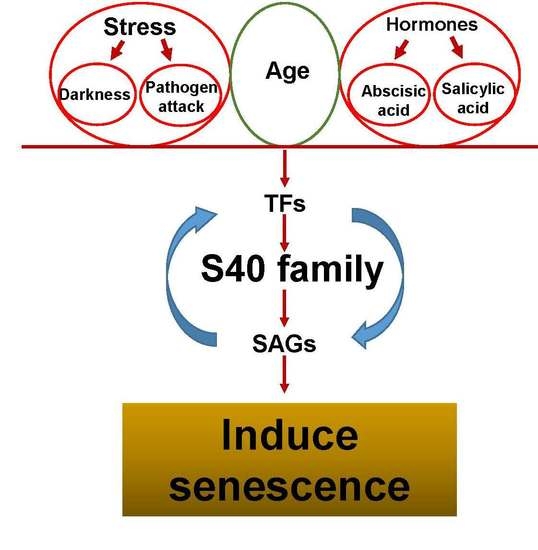
:1. Introduction
2. Phylogenetic Tree Construction
3. Alterations in S40 Genes Correspond to Age-Dependent as Well as Artificially-Induced Leaf Senescence
4. Subcellular Localization
5. Natural Leaf Senescence Is Delayed and SAGs Are Repressed in the S40 Mutant
6. Senescence-Related Expression of the S40 Gene Shows Regulation at the Transcriptional Level
7. Cis Elements Comparison in the Promoters of S40 Genes in Barley, Arabidopsis and Rice
8. The WHIRLY1 Protein Binds to HvS40 Promoters at the MII Motif Region
9. Conclusions
Conflicts of Interest
References
- Noodén, L. The Phenomena of Senescence and Aging; Food and Agriculture Organization: Rome, Italy, 1988. [Google Scholar]
- Matile, P. Chloroplast senescence. In Crop Photosynthesis: Spatial and Temporal Determinants; Elsevier: Amsterdam, The Netherlands, 1992; pp. 413–440. [Google Scholar]
- Kohl, S.; Hollmann, J.; Blattner, F.R.; Radchuk, V.; Andersch, F.; Steuernagel, B.; Schmutzer, T.; Scholz, U.; Krupinska, K.; Weber, H. A putative role for amino acid permeases in sink-source communication of barley tissues uncovered by RNA-seq. BMC Plant Biol. 2012, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, J.; Gregersen, P.L.; Krupinska, K. Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of field grown barley. J. Exp. Bot. 2014, 65, 3963–3973. [Google Scholar] [CrossRef] [PubMed]
- Pleuel, H.; Ojanpera, K.; Danielsson, H.; Sild, E.; Gelang, J.; Wallin, G. Effects of Ozone on Leaf Senescence in Spring Wheat—Possible Consequences for Grain Yield; Food and Agriculture Organization: Rome, Italy, 1997. [Google Scholar]
- Li, H.; Wang, S.; Li, B.; Tong, Y.; Yang, X.; Li, Z. Comparative study on physiological traits related with grain filling and photosynthesis of flag leaf in early aging and normal near—Isogenic lines of common wheat. Zuo Wu Xue Bao 2005, 32, 1642–1648. [Google Scholar]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Duvick, D.N. Genetic contributions to yield gains of US hybrid maize, 1930 to 1980. In Genetic Contributions to Yield Gains of Five Major Crop Plants; Crop Science Society of America and American Society of Agronomy: Madison, WI, USA, 1984; pp. 15–47. [Google Scholar]
- Tollenaar, M.; Wu, J. Yield improvement in temperate maize is attributable to greater stress tolerance. Crop Sci. 1999, 39, 1597–1604. [Google Scholar] [CrossRef]
- Egli, D.B. Seed-fill duration and yield of grain crops. Adv. Agron. 2004, 83, 243–279. [Google Scholar]
- Guo, Y.; Gan, S.-S. Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 2014, 65, 3901–3913. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Porc. Natl. Acad. Sic. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, C.S.; Halevy, A.H.; Reid, M.S. Roles of ethylene and 1-aminocyclopropane-1-carboxylic acid in pollination and wound-induced senescence of Petunia hybrida flowers. Physiol. Plant. 1984, 61, 643–648. [Google Scholar] [CrossRef]
- Becker, W.; Apel, K. Differences in gene expression between natural and artificially induced leaf senescence. Planta 1993, 189, 74–79. [Google Scholar] [CrossRef]
- Lutts, S.; Kinet, J.; Bouharmont, J. NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann. Bot. 1996, 78, 389–398. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; GAN, S.S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Gan, S. Molecular characteristics of leaf senescence. Plant Mol. Biol. 2003, 1, 1–17. [Google Scholar]
- Gan, S. Mitotic and postmitotic senescence in plants. Sci. Ageing Knowl. Environ. 2003. [Google Scholar] [CrossRef] [PubMed]
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Buchanan-Wollaston, V.; Ainsworth, C. Leaf senescence in Brassica napus: Cloning of senescence related genes by subtractive hybridisation. Plant Mol. Biol. 1997, 33, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, Y.; Breeze, E.; Koo, J.C.; Woo, H.R.; Ryu, J.S.; Park, D.H.; Beynon, J.; Tabrett, A.; Buchanan-Wollaston, V. Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 2007, 52, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tang, W.; Swain, J.D.; Green, A.L.; Jack, T.P.; Gan, S. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 2001, 126, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.-S.; Penfold, C.A.; Jenkins, D. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, U.; Jobst, J.; Kolb, D.; Rentsch, D. Senescence-related gene expression profiles of rosette leaves of Arabidopsis thaliana: Leaf age versus plant age. Plant Biol. 2004, 6, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flügge, U.-I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Holm, P.B. Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 2007, 5, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Parrott, D.L.; McInnerney, K.; Feller, U.; Fischer, A.M. Steam-girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence-associated genes. New Phytol. 2007, 176, 56–69. [Google Scholar] [CrossRef] [PubMed]
- De Michele, R.; Formentin, E.; Todesco, M.; Toppo, S.; Carimi, F.; Zottini, M.; Barizza, E.; Ferrarini, A.; Delledonne, M.; Fontana, P. Transcriptome analysis of Medicago truncatula leaf senescence: Similarities and differences in metabolic and transcriptional regulations as compared with Arabidopsis, nodule senescence and nitric oxide signalling. New Phytol. 2009, 181, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhou, Y.; Zhou, G.; Ye, R.; Zhao, L.; Li, X.; Lin, Y. Identification of early senescence-associated genes in rice flag leaves. Plant Mol. Biol. 2008, 67, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.; Keskitalo, J.; Sjödin, A.; Bhalerao, R.; Sterky, F.; Wissel, K.; Tandre, K.; Aspeborg, H.; Moyle, R.; Ohmiya, Y. A transcriptional timetable of autumn senescence. Genome Biol. 2004, 5, R24. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y. Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 2013, 82, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, Y.; Tanaka, T.; Ogiwara, I.; Kanekatsu, M.; van Doorn, W.G.; Yamada, T. Expression of an AtNAP gene homolog in senescing morning glory (Ipomoea nil) petals of two cultivars with a different flower life span. J. Plant Physiol. 2014, 171, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-J.; Wei, Q.; Kuai, B.-K. Cloning and characterization of the AtNAP orthologous in Festuca arundinacea. J. Fudan Univ. (Nat. Sci.) 2010, 49, 544–551. [Google Scholar]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qiu, K.; Kuai, B.; Ding, Y. Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Physiol. Plant. 2011, 142, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.-R.; Zhang, R.-K.; Chen, X.; Wu, X.-J.; Ming, F. Characterization of OsNAP from Oryza sativa L. and its application in molecular breeding. J. Fudan Univ. (Nat. Sci.) 2012, 51, 507–514. [Google Scholar]
- Zhang, Y.; Cao, Y.; Shao, Q.; Wang, L.; Wang, H.; Li, J.; Li, H. Regulating Effect of ZmNAP Gene on Anti-senescence and Yield Traits of Maize. J. Henan Agric. Sci. 2012, 41, 19–24. [Google Scholar]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 2011, 31, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.; Zhou, X.; Song, Y.; Yu, D. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. 2009, 58, 181–190. [Google Scholar] [CrossRef]
- Han, M.; Kim, C.-Y.; Lee, J.; Lee, S.-K.; Jeon, J.-S. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 2014, 37, 532. [Google Scholar] [CrossRef] [PubMed]
- Ricachenevsky, F.K.; Sperotto, R.A.; Menguer, P.K.; Fett, J.P. Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol. Biol. Rep. 2010, 37, 3735–3745. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence-and defence-related processes. Plant J. 2001, 28, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Kilbienski, I. Untersuchungen zur Funktion und Regulation des Seneszenzassoziierten HvS40-Genes der Gerste (Hordeum vulgare L.) und HvS40 Verwandter Gene von Arabidopsis Thaliana; Christian-Albrechts Universität Kiel: Kiel, Germany, 2007. [Google Scholar]
- Fischer-Kilbienski, I.; Miao, Y.; Roitsch, T.; Zschiesche, W.; Humbeck, K.; Krupinska, K. Nuclear targeted AtS40 modulates senescence associated gene expression in Arabidopsis thaliana during natural development and in darkness. Plant Mol. Biol. 2010, 73, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Humbeck, K.; Quast, S.; Krupinska, K. Functional and molecular changes in the photosynthetic apparatus during senescence of flag leaves from field-grown barley plants. Plant Cell Environ. 1996, 19, 337–344. [Google Scholar] [CrossRef]
- Falk, J.; Krauß, N.; Dähnhardt, D.; Krupinska, K. The senescence associated gene of barley encoding 4-hydroxyphenylpyruvate dioxygenase is expressed during oxidative stress. J. Plant Physiol. 2002, 159, 1245–1253. [Google Scholar] [CrossRef]
- Haussuhl, K. Charakterisierung der Blattseneszenz von Hordeum vulgare L. Unter Besonderer Berucksichtigung des Gens HvS40; Christian-Albrechts-Universität zu Kiel: Kiel, Germany, 1998. [Google Scholar]
- Krupinska, K.; Haussühl, K.; Schäfer, A.; van der Kooij, T.A.; Leckband, G.; Lörz, H.; Falk, J. A novel nucleus-targeted protein is expressed in barley leaves during senescence and pathogen infection. Plant Physiol. 2002, 130, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Krupinska, K.; Dähnhardt, D.; Fischer-Kilbienski, I.; Kucharewicz, W.; Scharrenberg, C.; Trösch, M.; Buck, F. Identification of WHIRLY1 as a factor binding to the promoter of the stress-and senescence-associated gene HvS40. Plant Growth Regul. 2014, 33, 91–105. [Google Scholar] [CrossRef]
- Kleber-Janke, T.; Krupinska, K. Isolation of cDNA clones for genes showing enhanced expression in barley leaves during dark-induced senescence as well as during senescence under field conditions. Planta 1997, 203, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Lee, S.Y.; Chung, I.K.; Lee, C.-H.; Nam, H.G. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 1996, 30, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Krause, K.; Falk, J.; Humbeck, K.; Krupinska, K. Responses of the transcriptional apparatus of barley chloroplasts to a prolonged dark period and to subsequent reillumination. Physiol. Plant. 1998, 104, 143–152. [Google Scholar] [CrossRef]
- Scharrenberg, C. Untersuchungen zur Transkriptionellen Kontrolle der Genexpression Während der Blattseneszenz der Gerste (Hordeum vulgare L.); Christian-Albrechts Universität Kiel: Kiel, Germany, 2002. [Google Scholar]
- Rushton, P.J.; Somssich, I.E. Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1998, 1, 311–315. [Google Scholar] [CrossRef]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Desveaux, D.; Després, C.; Joyeux, A.; Subramaniam, R.; Brisson, N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 2000, 12, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Desveaux, D.; Subramaniam, R.; Després, C.; Mess, J.-N.; Lévesque, C.; Fobert, P.R.; Dangl, J.L.; Brisson, N. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 2004, 6, 229–240. [Google Scholar] [CrossRef]
- Robatzek, S.; Somssich, I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Bonetta, D.; McCourt, P. Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 1998, 3, 231–235. [Google Scholar] [CrossRef]
- Grill, E.; Himmelbach, A. ABA signal transduction. Curr. Opin. Plant Biol. 1998, 1, 412–418. [Google Scholar] [CrossRef]
- Guiltinan, M.J.; Marcotte, W.R., Jr.; Quatrano, R.S. A plant leucine zipper protein that recognizes an abscisic acid response element. Science 1990, 250, 267. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 2000, 21, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Schmelzer, E.; Hahlbrock, K.; Somssich, I.E. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef] [PubMed]
- Marè, C.; Mazzucotelli, E.; Crosatti, C.; Francia, E.; Stanca, A.M.; Cattivelli, L. Hv-WRKY38: A new transcription factor involved in cold-and drought-response in barley. Plant Mol. Biol. 2004, 55, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef] [PubMed]





| S40 Genes | Age Dependent Senescence | Salt Stress | Drought Stress | Cold Stress |
|---|---|---|---|---|
| OsS40-1 | Slightly increased | Increased | No change | No change |
| OsS40-2 | No change | No change | No change | No change |
| OsS40-3 | No change | No change | No change | No change |
| OsS40-4 | Increased | Increased | No change | No change |
| OsS40-5 | No change | No change | Increased | No change |
| OsS40-6 | No change | Increased | Increased | No change |
| OsS40-7 | Increased | Increased | Increased | No change |
| OsS40-8 | No change | No change | No change | No change |
| OsS40-9 | No change | Increased | Increased | No change |
| OsS40-10 | No change | No change | No change | No change |
| OsS40-11 | No change | No change | No change | No change |
| OsS40-12 | No change | No change | No change | No change |
| OsS40-13 | Slightly increased | Increased | No change | No change |
| OsS40-14 | Increased | No change | Increased | No change |
| OsS40-15 | Increased | Increased | No change | No change |
| OsS40-16 | Slightly increased | Increased | No change | No change |
| Loci Number in Genome | Gene Name | Yloc (http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi) | WoLF PSORT (https://wolfpsort.hgc.jp/) | DISTILL 2.0 (http://distillf.ucd.ie/distill/) | LOCALI-ZER 1.0 (http://localizer.csiro.au/) | BaCelLo (http://gpcr.biocomp.unibo.it/bacello/pred.htm) | Euk-mPLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/euk-?multi-2/) |
|---|---|---|---|---|---|---|---|
| Loc_Hv FI496079 | HvS40—Nucleus (Krupinska et al. 2002) [54] | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Mitochondria. Nucleus |
| LOC_Os05g45450 | OsS40-1 | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus |
| LOC_Os05g44260 | OsS40-2 | Nucleus | Mitochondria | Nucleus | Nucleus | Nucleus | Extra cell |
| LOC_Os10g27350 | OsS40-3 | Nucleus | Chloroplast | Chloroplast | ------ | Nucleus | Nucleus |
| LOC_Os03g02280 | OsS40-4 | Nucleus | Chloroplast | Chloroplast | ------ | Secretory | Extra cell |
| LOC_Os04g45834 | OsS40-5 | Nucleus | Chloroplast | Chloroplast | Nucleus | Nucleus | Nucleus, Extra cell |
| LOC_Os04g33760 | OsS40-6 | Nucleus | Nucleus | Chloroplast | Nucleus | Nucleus | Nucleus, Extra cell |
| LOC_Os01g52730 | OsS40-7 | Nucleus | Nucleus | Chloroplast | ------- | Nucleus | Extra cell |
| LOC_Os10g33990 | OsS40-8 | Cytoplasm | Chloroplast | Chloroplast | Nucleus | Nucleus | Extra cell |
| LOC_Os01g64300 | OsS40-9 | Nucleus | Nucleus | Chloroplast | ------- | Nucleus | Extra cell |
| LOC_Os01g52740 | OsS40-10 | Cytoplasm | Nucleus | Chloroplast | Nucleus | Nucleus | Nucleus, Extra cell |
| LOC_Os12g05980 | OsS40-11 | Mitochondria | Chloroplast | Chloroplast | Nucleus | Nucleus | Extra cell |
| LOC_Os11g05600 | OsS40-12 | Chloroplast | Chloroplast | Chloroplast | Nucleus | Nucleus | Extra cell |
| LOC_Os04g43990 | OsS40-13 | Chloroplast | Mitochondria | Chloroplast | --------- | Chloroplast | Extra cell |
| LOC_Os05g45440 | OsS40-14 | Nucleus | Cytoplasm | Chloroplast | Nucleus | Nucleus | Nucleus |
| LOC_Os07g33270 | OsS40-15 | Nucleus | Mitochondria | Chloroplast | --------- | Nucleus | Extra cell |
| LOC_Os07g32810 | OsS40-16 | Nucleus | Nucleus | Chloroplast | ---------- | Nucleus | Extra cell |
| Cis-Elements | HvS40 | HvSDI | HvSD8 | At2g 34340 | At1g 29640 | At5g 45630 | AtS4-3 | OsS40-1 | OsS40-2 | OsS40-7 | OsS40-14 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| W-box | 4 | 4 | 5 | 5 | 6 | 5 | 2 | 2 | 2 | 5 | 3 |
| ERE | 2 | 1 | |||||||||
| MYB | 19 | 3 | 2 | 1 | 2 | 6 | 7 | 5 | 3 | 5 | 6 |
| LREs | 13 | 11 | 7 | 2 | 1 | ||||||
| MYC | 8 | 2 | 2 | 6 | 4 | 10 | 8 | 18 | 6 | 4 | 8 |
| ABRE | 6 | 5 | 4 | 4 | 15 | 15 | 6 | 4 | 8 | 4 | |
| Dof | 5 | 19 | 7 | 11 | 18 | 12 | 26 | 4 | 12 | 8 | 5 |
| PRE | 5 | 15 | 11 | ||||||||
| CGCG box | 3 | 3 | 3 | 8 | 26 | 14 | 20 | ||||
| SURE | 2 | 4 | 6 | 5 | 1 | 5 | 3 | 1 | 1 | ||
| DRE/CRT | 2 | 4 | 3 | 1 | 1 | 2 | 2 | 1 | |||
| LTR | 2 | 2 | 3 | 1 | 1 | 1 | 3 | 2 | 3 | ||
| ARF | 2 | 3 | 2 | 1 | 1 | ||||||
| DPBFCOREDCDC3 | 2 | 1 | 3 | 4 | 6 | 2 | 1 | 3 | 2 | ||
| Pyrimidine box | 1 | 3 | 2 | 1 | 2 | 1 | 2 | 1 | 3 | 1 | |
| G-box plus G | 1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jehanzeb, M.; Zheng, X.; Miao, Y. The Role of the S40 Gene Family in Leaf Senescence. Int. J. Mol. Sci. 2017, 18, 2152. https://doi.org/10.3390/ijms18102152
Jehanzeb M, Zheng X, Miao Y. The Role of the S40 Gene Family in Leaf Senescence. International Journal of Molecular Sciences. 2017; 18(10):2152. https://doi.org/10.3390/ijms18102152
Chicago/Turabian StyleJehanzeb, Muhammad, Xiangzi Zheng, and Ying Miao. 2017. "The Role of the S40 Gene Family in Leaf Senescence" International Journal of Molecular Sciences 18, no. 10: 2152. https://doi.org/10.3390/ijms18102152
APA StyleJehanzeb, M., Zheng, X., & Miao, Y. (2017). The Role of the S40 Gene Family in Leaf Senescence. International Journal of Molecular Sciences, 18(10), 2152. https://doi.org/10.3390/ijms18102152