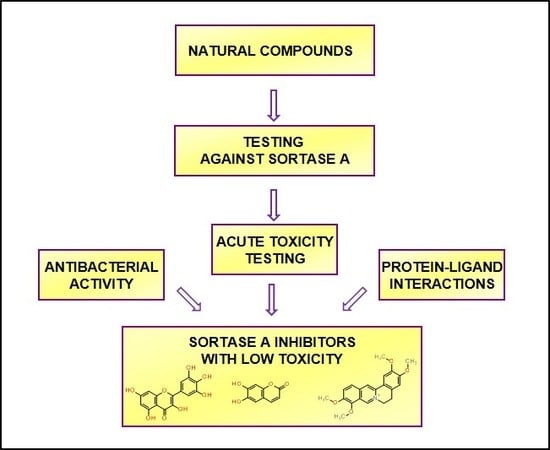
Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors
Abstract
:1. Introduction
2. Results
2.1. Sortase A Activity Assay
2.2. Protein-Ligand Interactions
2.3. Antibacterial Activity
2.4. Acute Toxicity Assessment Using Daphnia Magna
3. Discussion
4. Materials and Methods
4.1. Sortase A Activity Assay
4.2. Molecular Docking
4.2.1. Protein and Ligand Preparation
4.2.2. Protein-Ligand Interaction Prediction
4.3. Antibacterial Assay
4.3.1. Well Diffusion Test
4.3.2. Microdilution Test
4.4. Acute Toxicity Assessment Using Daphnia Magna
4.5. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CFU | colony-forming unit |
| CI | confidence interval |
| DMSO | dimethyl sulfoxide |
| FRET | fluorescence resonance energy transfer |
| HMB | 4-hydroxymercuribenzoic acid |
| IC50 | half maximal inhibitory concentration |
| LC50 | median lethal dose |
| MBH | Mueller-Hinton broth |
| MDR | multi-drug resistant |
| MIC | minimum inhibitory concentration |
| SAR | structure-activity relationships |
| SrtA | sortase A |
References
- Haaber, J.; Penadés, J.R.; Ingmer, H. Transfer of Antibiotic Resistance in Staphylococcus aureus. Trends Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Lebreton, F.; van Schaik, W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 2013, 16, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Kaczorek, E.; Małaczewska, J.; Wójcik, R.; Rękawek, W.; Siwicki, A.K. Phenotypic and genotypic antimicrobial susceptibility pattern of Streptococcus spp. isolated from cases of clinical mastitis in dairy cattle in Poland. J. Dairy Sci. 2017, 100, 6442–6453. [Google Scholar] [CrossRef] [PubMed]
- The World Health Organization (WHO). WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Zhang, Y.; Zhang, J.; Chen, W.; Angsantikul, P.; Spiekermann, K.A.; Fang, R.H.; Gao, W.; Zhang, L. Erythrocyte membrane-coated nanogel for combinatorial antivirulence and responsive antimicrobial delivery against Staphylococcus aureus infection. J. Control. Release 2017, 263, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Clewell, D.B.; Ike, Y.; Shankar, N. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Saichek, N.R.; Cox, C.R.; Kim, S.; Harrington, P.B.; Stambach, N.R.; Voorhees, K.J. Strain-level Staphylococcus differentiation by CeO2-metal oxide laser ionization mass spectrometry fatty acid profiling. BMC Microbiol. 2016, 16, 72. [Google Scholar] [CrossRef] [PubMed]
- McMahon, R.M.; Premkumar, L.; Martin, J.L. Four structural subclasses of the antivirulence drug target disulfide oxidoreductase DsbA provide a platform for design of subclass-specific inhibitors. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Dickey, S.W.; Cheung, G.Y.C.; Otto, M. Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017, 16, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Dong, H.; Murfin, K.E.; Feng, C.; Wu, S.; Zheng, B. Active site analysis of sortase A from Staphylococcus simulans indicates function in cleavage of putative cell wall proteins. Biochem. Biophys. Res. Commun. 2016, 478, 1653–1659. [Google Scholar] [CrossRef] [PubMed]
- Antos, J.M.; Ingram, J.; Fang, T.; Pishesha, N.; Truttmann, M.C.; Ploegh, H.L. Site-Specific Protein Labeling via Sortase-Mediated Transpeptidation. Curr. Protoc. Protein Sci. 2017, 15.3.1–15.3.19. [Google Scholar] [CrossRef]
- Shrestha, P.; Wereszczynski, J. Discerning the catalytic mechanism of Staphylococcus aureus sortase A with QM/MM free energy calculations. J. Mol. Graph. Model. 2016, 67, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Marraffini, L.A.; Dedent, A.C.; Schneewind, O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 192–221. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-Y.; Kim, C.-K.; Ahn, C.-H.; Kim, H.; Shin, J.; Oh, K.-B. Flavonoid glycosides inhibit sortase A and sortase A-mediated aggregation of Streptococcus mutans, an oral bacterium responsible for human dental caries. J. Microbiol. Biotechnol. 2016, 26, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Orhan, D.D.; Özçelik, B.; Özgen, S.; Ergun, F. Antibacterial, antifungal, and antiviral activities of some flavonoids. Microbiol. Res. 2010, 165, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Kim, Y.-G.; Cho, H.S.; Ryu, S.Y.; Cho, M.H.; Lee, J. Coumarins reduce biofilm formation and the virulence of Escherichia coli O157:H7. Phytomedicine 2014, 21, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Comini, L.R.; Núñez Montoya, S.C.; Páez, P.L.; Argüello, G.A.; Albesa, I.; Cabrera, J.L. Antibacterial activity of anthraquinone derivatives from Heterophyllaea pustulata (Rubiaceae). J. Photochem. Photobiol. B Biol. 2011, 102, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Cushnie, B.; Lamb, A.J. Alkaloids: An overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents 2014, 44, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.; Zanfirescu, A.; Olaru, O.T.; Nicorescu, I.M.; Nitulescu, G.M.; Margina, D. Structural analysis of sortase A inhibitors. Molecules 2016, 21, 1591. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Bice, T.W.; Ton-That, H.; Schneewind, O.; Narayana, S.V.L. Crystal structures of Staphylococcus aureus Sortase A and its substrate complex. J. Biol. Chem. 2004, 279, 31383–31389. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, F.; Bi, C.; Wang, L.; Zhong, X.; Cai, H.; Deng, X.; Niu, X.; Wang, D. Quercitrin, an Inhibitor of Sortase A, Interferes with the Adhesion of Staphylococcal aureus. Molecules 2015, 20, 6533–6543. [Google Scholar] [CrossRef] [PubMed]
- Ang, S.S.K.; Im, J.K.; Ee, T.L. Flavonols Inhibit Sortases and Sortase-Mediated Staphylococcus aureus Clumping to Fibrinogen. Biol. Pharm. Bull. 2006, 29, 1751. [Google Scholar]
- Suree, N.; Yi, S.W.; Thieu, W.; Marohn, M.; Damoiseaux, R.; Chan, A.; Jung, M.E.; Clubb, R.T. Discovery and structure-activity relationship analysis of Staphylococcus aureus sortase A inhibitors. Bioorg. Med. Chem. 2009, 17, 7174–7185. [Google Scholar] [CrossRef] [PubMed]
- Lippert, T.; Rarey, M. Fast automated placement of polar hydrogen atoms in protein-ligand complexes. J. Cheminform. 2009, 1. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2011, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.P.; Kaur, A.; Singh, N.; Nim, L.; Shevkani, K.; Kaur, H.; Arora, D.S. In vitro antioxidant and antimicrobial properties of jambolan (Syzygium cumini) fruit polyphenols. LWT Food Sci. Technol. 2016, 65, 1025–1030. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI: Wayne, PA, USA, 2016; CLSI Supplement M100S. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved St andard—Ninth Edition; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2012; Volume 32, ISBN 1562387839. [Google Scholar]
- Pirvu, L.; Nicorescu, I.; Hlevca, C.; Albu, B.; Nicorescu, V. Burdock (Arctium lappa) Leaf Extracts Increase the In Vitro Antimicrobial Efficacy of Common Antibiotics on Gram-positive and Gram-negative Bacteria. Open Chem. 2017, 15, 92. [Google Scholar] [CrossRef]
- Andrews, J.M.; Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Olaru, O.T.; Venables, L.; Van De Venter, M.; Nitulescu, G.M.; Margina, D.; Spndidos, D.A.; Tsatsakis, A.M. Anticancer potential of selected Fallopia Adans species. Oncol. Lett. 2015, 10, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Socea, L.I.; Socea, B.; Sarame, G.; Barbuceanu, S.F.; Draghici, C.; Constantin, V.D.; Olaru, O.T. Synthesis and cytotoxicity evaluation of new 5H-dibenzo[a,d] [7]annulen-5-yl acetylhydrazones. Rev. Chim. 2015, 66, 1122–1127. [Google Scholar]
- Pahonţu, E.; Ilieş, D.C.; Shova, S.; Oprean, C.; Pǎunescu, V.; Olaru, O.T.; Rǎdulescu, F.Ş.; Gulea, A.; Roşu, T.; Drǎgǎnescu, D. Synthesis, characterization, antimicrobial and antiproliferative activity evaluation of Cu(II), Co(II), Zn(II), Ni(II) and Pt(II) complexes with isoniazid-derived compound. Molecules 2017, 22, 650. [Google Scholar] [CrossRef] [PubMed]
- OECD Test No. 202: Daphnia sp. Acute Immobilisation Test; OECD Test Guidelines including Chemical Testing and Assessment Section 2; Organization for Economic Co-operation and Development: Paris, France, 2004; pp. 1–12.










| Compound | Concentration (μM) | Inhibition (%) | IC50 (μM) | 95% CI of IC50 (μM) |
|---|---|---|---|---|
| Myricetin | 100 | 91.96 | 4.63 | 0.19–111.0 |
| Esculetin | 250 | 83.57 | 36.16 | 3.18–410.4 |
| Palmatine chloride | 500 | 85.29 | 52.84 | 3.46–805.7 |
| Rutin trihydrate | 500 | 28.71 | NC * | NC * |
| Podophyllotoxin | 50 | 26.40 | NC * | NC * |
| Troxerutin | 500 | 23.50 | NC * | NC * |
| Quinic acid | 500 | 19.56 | NC * | NC * |
| Piperine | 10 | 14.88 | NT ** | NC * |
| Emodin | 100 | 10.14 | NC * | NC * |
| Genistein | 500 | 9.63 | NC * | NC * |
| Quercetin | 10 | 8.55 | NT ** | NC * |
| Rhein | 10 | 7.01 | NT ** | NC * |
| Artemisinin | 50 | 5.32 | NC * | NC * |
| Umbelliferone | 250 | 4.04 | NC * | NC * |
| HMB | 5 | 93.87 | 0.114 | 0.02–0.50 |
| Compound | S. aureus ATCC 6538 | S. aureus ATCC 25923 | S. epidermidis ATCC 12228 | E. faecalis ATCC 29212 | B. cereus ATCC 11778 | E. coli ATCC 8739 | E. coli ATCC 35218 | P. mirabilis ATCC 29245 |
|---|---|---|---|---|---|---|---|---|
| Diameters of inhibition zone (mm), including diameter of well | ||||||||
| Gentamicin (10 μg) | 17 | 21 | 29 | 12 | 24 | 21 | 20 | 20 |
| Emodin (13.5 μg) | 6 | 20 | 14 | 6 | 12 | 6 | 6 | 6 |
| Rhein (14.2 μg) | 6 | 12 | 30 | 6 | 15 | 6 | 6 | 6 |
| Compound | S. aureus ATCC 25923 | S. epidermidis ATCC 12228 | B. cereus ATCC 11778 |
|---|---|---|---|
| Emodin | 8 | 8 | 16 |
| Rhein | 64 | 16 | 32 |
| Gentamicin | 2 | 2 | 4 |
| Compound | LC50 (μM) | 95% CI of LC50 (μM) | ||
|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | |
| Piperine | 144.60 | 3.45 | 82.26–254.1 | 1.89–6.27 |
| Podophyllotoxin | NC * | 6.47 | NC * | 4.01–10.46 |
| Chrysin | NC * | 17.68 | NC * | 7.73–40.44 |
| Emodin | 34.67 | 24.15 | 25.67–46.83 | 16.70–34.92 |
| Umbelliferone | NC * | 104.90 | NC * | 21.81–504.60 |
| Potassium dichromate | 16.39 | 0.62 | 4.15–5.59 | NC * |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nitulescu, G.; Nicorescu, I.M.; Olaru, O.T.; Ungurianu, A.; Mihai, D.P.; Zanfirescu, A.; Nitulescu, G.M.; Margina, D. Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors. Int. J. Mol. Sci. 2017, 18, 2217. https://doi.org/10.3390/ijms18102217
Nitulescu G, Nicorescu IM, Olaru OT, Ungurianu A, Mihai DP, Zanfirescu A, Nitulescu GM, Margina D. Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors. International Journal of Molecular Sciences. 2017; 18(10):2217. https://doi.org/10.3390/ijms18102217
Chicago/Turabian StyleNitulescu, Georgiana, Isabela Madalina Nicorescu, Octavian Tudorel Olaru, Anca Ungurianu, Dragos Paul Mihai, Anca Zanfirescu, George Mihai Nitulescu, and Denisa Margina. 2017. "Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors" International Journal of Molecular Sciences 18, no. 10: 2217. https://doi.org/10.3390/ijms18102217
APA StyleNitulescu, G., Nicorescu, I. M., Olaru, O. T., Ungurianu, A., Mihai, D. P., Zanfirescu, A., Nitulescu, G. M., & Margina, D. (2017). Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors. International Journal of Molecular Sciences, 18(10), 2217. https://doi.org/10.3390/ijms18102217