PeaTAR1B: Characterization of a Second Type 1 Tyramine Receptor of the American Cockroach, Periplaneta americana
Abstract
:1. Introduction
2. Results
2.1. Cloning of Peatar1b cDNA and Structural Properties of PeaTAR1B
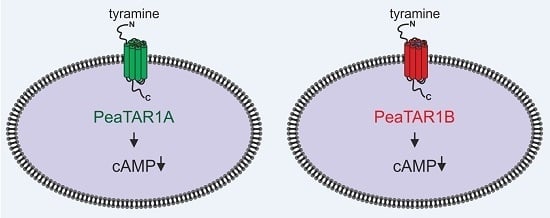
2.2. Functional and Pharmacological Properties of PeaTAR1B
3. Discussion
3.1. Molecular Features of the PeaTAR1B Receptor
3.2. Pharmacological Properties of PeaTAR1B
3.3. Expression Pattern
4. Materials and Methods
4.1. Cloning of the Peatar1b cDNA
4.2. Multiple Sequence Alignments and Phylogenetic Analysis
4.3. Construction of Expression Vectors
4.4. Functional Expression in Mammalian Cell Lines
4.5. Functional Characterization of the PeaTAR1B Receptor
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| cAMP | cyclic adenosine monophosphate (3′,5′-cyclic adenosine monophosphate) |
| GPCR | G protein-coupled receptor |
| IBMX | 3-isobutyl-1-methylxanthine |
| ID | identity |
| NCBI | National Center for Biotechnology Information |
| PKA | protein kinase A (AMP-dependent protein kinase) |
| PKC | protein kinase C |
| S | similarity |
| TAAR | trace amine-associated receptor |
| TM | transmembrane domain |
References
- Huber, I.; Masler, E.P.; Rao, B.R. Cockroaches as Models for Neurobiology: Applications in Biomedical Research, 1st ed.; CRC Press: Boca Raton, FL, USA, 1990. [Google Scholar]
- Walz, B.; Baumann, O.; Krach, C.; Baumann, A.; Blenau, W. The aminergic control of cockroach salivary glands. Arch. Insect Biochem. Physiol. 2006, 62, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, M.; Dąbrowski, M.; de Lima, M.E. Nervous system of Periplaneta americana cockroach as a model in toxinological studies: A short historical and actual view. J. Toxicol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Camhi, J.M. Escape behavior in the cockroach: Distributed neural processing. Experientia 1988, 44, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Domenici, P.; Booth, D.; Blagburn, J.M.; Bacon, J.P. Cockroaches keep predators guessing by using preferred escape trajectories. Curr. Biol. 2008, 18, 1792–1796. [Google Scholar] [CrossRef] [PubMed]
- Helfrich-Förster, C. The circadian clock in the brain: A structural and functional comparison between mammals and insects. J. Comp. Physiol. A 2004, 190, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Matsumoto, T.; Ichihara, N.; Sakai, T.; Satake, H.; Watari, Y.; Takeda, M. The pars intercerebralis as a modulator of locomotor rhythms and feeding in the American cockroach, Periplaneta americana. Physiol. Behav. 2009, 96, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Wegener, C.; Herbert, Z.; Eckert, M.; Predel, R. The periviscerokinin (PVK) peptide family in insects: Evidence for the inclusion of CAP2b as a PVK family member. Peptides 2002, 23, 605–611. [Google Scholar] [CrossRef]
- Neupert, S.; Fusca, D.; Schachtner, J.; Kloppenburg, P.; Predel, R. Toward a single-cell-based analysis of neuropeptide expression in Periplaneta americana antennal lobe neurons. J. Comp. Neurol. 2012, 520, 694–716. [Google Scholar] [CrossRef] [PubMed]
- Arendt, A.; Neupert, S.; Schendzielorz, J.; Predel, R.; Stengl, M. The neuropeptide SIFamide in the brain of three cockroach species. J. Comp. Neurol. 2016, 524, 1337–1360. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Mizunami, M. Classical conditioning of activities of salivary neurones in the cockroach. J. Exp. Biol. 2006, 209, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Mizunami, M. Pavlov’s cockroach: Classical conditioning of salivation in an insect. PLoS ONE 2007, 2, e529. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Sitcom, C.; Kuramochi, T.; Nishino, H.; Mizunami, M. Salivary conditioning with antennal gustatory unconditioned stimulus in an insect. Neurobiol. Learn. Mem. 2008, 90, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, C.S.; Matsumoto, Y.; Watanabe, H.; Nishino, H.; Mizunami, M. Context-dependent olfactory learning monitored by activities of salivary neurons in cockroaches. Neurobiol. Learn. Mem. 2012, 97, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Hosono, S.; Matsumoto, Y.; Mizunami, M. Interaction of inhibitory and facilitatory effects of conditioning trials on long-term memory formation. Learn. Mem. 2016, 23, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Marg, S.; Walz, B.; Blenau, W. The effects of dopamine receptor agonists and antagonists on the secretory rate of cockroach (Periplaneta americana) salivary glands. J. Insect Physiol. 2004, 50, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Rietdorf, K.; Blenau, W.; Walz, B. Protein secretion in cockroach salivary glands requires an increase in intracellular cAMP and Ca2+ concentrations. J. Insect Physiol. 2005, 51, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Troppmann, B.; Walz, B.; Blenau, W. Pharmacology of serotonin-induced salivary secretion in Periplaneta americana. J. Insect Physiol. 2007, 53, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Hille, C.; Walz, B. A vacuolar-type H+-ATPase and a Na+/H+ exchanger contribute to intracellular pH regulation in cockroach salivary ducts. J. Exp. Biol. 2007, 210, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Rotte, C.; Witte, J.; Blenau, W.; Baumann, O.; Walz, B. Source, topography and excitatory effects of GABAergic innervation in cockroach salivary glands. J. Exp. Biol. 2009, 212, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Khan, A.M.; Baqui, M.A.; Muhibullah, M. Evaluation of the common cockroach Periplaneta americana (L.) as carrier of medically important bacteria. J. Commun. Dis. 1992, 24, 206–210. [Google Scholar] [PubMed]
- García, F.; Notario, M.J.; Cabanás, J.M.; Jordano, R.; Medina, L.M. Incidence of bacteria of public health interest carried by cockroaches in different food-related environments. J. Med. Entomol. 2012, 49, 1481–1484. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Vellody, D.; Homburger, H.; Yunginger, J.W. Cockroach cause of allergic asthma. Its specificity and immunologic profile. J. Allergy Clin. Immunol. 1979, 63, 80–86. [Google Scholar] [CrossRef]
- Arruda, L.K.; Vailes, L.D.; Ferriani, V.P.; Santos, A.B.; Pomés, A.; Chapman, M.D. Cockroach allergens and asthma. J. Allergy Clin. Immunol. 2001, 107, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Sookrung, N.; Chaicumpa, W. A revisit to cockroach allergens. Asian Pac. J. Allergy Immunol. 2010, 28, 95–106. [Google Scholar] [PubMed]
- Etan, E. Insecticidal activity of essential oils: Octopaminergic sites of action. Comp. Biochem. Physiol. C 2001, 130, 325–337. [Google Scholar]
- Enan, E.E. Molecular and pharmacological analysis of an octopamine receptor from American cockroach and fruit fly in response to plant essential oils. Arch. Insect Biochem. Physiol. 2005, 59, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Enan, E.E. Molecular response of Drosophila melanogaster tyramine receptor cascade to plant essential oils. Insect Biochem. Mol. Biol. 2005, 35, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Price, D.N.; Berry, M.S. Comparison of effects of octopamine and insecticidal essential oils on activity in the nerve cord, foregut, and dorsal unpaired median neurons of cockroaches. J. Insect Physiol. 2006, 52, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.D.; Gee, J.D. Action of formamidine pesticides on octopamine receptors. Nature 1980, 287, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Gole, J.W.; Orr, G.L.; Downer, R.G. Interaction of formamidines with octopamine-sensitive adenylate cyclase receptor in the nerve cord of Periplaneta americana L. Life Sci. 1983, 32, 2939–2947. [Google Scholar] [CrossRef]
- Downer, R.G.H.; Gole, J.W.D.; Orr, G.L. Interaction of formamidines with octopamine-, dopamine-, and 5-hydroxytryptamine-sensitive adenylate cyclase in the nerve cord of Periplaneta americana. Pestic. Sci. 1985, 16, 472–478. [Google Scholar] [CrossRef]
- Robb, S.; Cheek, T.R.; Hannan, F.L.; Hall, L.M.; Midgley, J.M.; Evans, P.D. Agonist-specific coupling of a cloned Drosophila octopamine/tyramine receptor to multiple second messenger systems. EMBO J. 1994, 13, 1325–1330. [Google Scholar] [PubMed]
- Huang, J.; Hamasaki, T.; Ozoe, Y. Pharmacological characterization of a Bombyx mori α-adrenergic-like octopamine receptor stably expressed in a mammalian cell line. Arch. Insect Biochem. Physiol. 2010, 73, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Kita, T.; Hayashi, T.; Ohtani, T.; Takao, H.; Takasu, H.; Liu, G.; Ohta, H.; Ozoe, F.; Ozoe, Y. Amitraz and its metabolite differentially activate α- and β-adrenergic-like octopamine receptors. Pest Manag. Sci. 2017, 73, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Regnault-Roger, C.; Vincent, C.; Arnason, J.T. Essential oils in insect control: Low-risk products in a high-stakes world. Annu. Rev. Entomol. 2012, 57, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Blenau, W.; Rademacher, E.; Baumann, A. Plant essential oils and formamidines as insecticides/acaricides: What are the molecular targets? Apidologie 2012, 43, 334–347. [Google Scholar] [CrossRef]
- Casida, J.E.; Durkin, K.A. Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annu. Rev. Entomol. 2013, 58, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Ozoe, Y. Molecular signalling, pharmacology, and physiology of octopamine and tyramine receptors as potential insect pest control targets. Adv. Insect Physiol. 2014, 46, 73–166. [Google Scholar] [CrossRef]
- Lange, A.B. Tyramine: From octopamine precursor to neuroactive chemical in insects. Gen. Comp. Endocrinol. 2009, 162, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, H.; Vleugels, R.; Martial, E.; Badisco, L.; Pflüger, H.J.; Blenau, W.; Vanden Broeck, J. The role of octopamine in locusts and other arthropods. J. Insect Physiol. 2010, 56, 854–867. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Guo, X.; Lei, H.; Li, T.; Hao, S.; Kang, L. Octopamine and tyramine respectively regulate attractive and repulsive behavior in locust phase changes. Sci. Rep. 2015, 5, 8036. [Google Scholar] [CrossRef] [PubMed]
- Blenau, W.; Baumann, A. Octopaminergic and tyraminergic signaling in the honeybee (Apis mellifera) brain: Behavioral, pharmacological, and molecular aspects. In Trace Amines and Neurological Disorders. Potential Mechanisms and Risk Factors, 1st ed.; Farooqui, T., Farooqui, A.A., Eds.; Academic Press: Oxford, UK, 2016; pp. 203–220. ISBN 9780128036037. [Google Scholar]
- Huang, J.; Liu, W.; Qi, Y.X.; Luo, J.; Montell, C. Neuromodulation of courtship drive through tyramine-responsive neurons in the Drosophila brain. Curr. Biol. 2016, 26, 2246–2256. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hoffmann, J.; Li, Y.; Stephano, F.; Bruchhaus, I.; Fink, C.; Roeder, T. Octopamine controls starvation resistance, life span and metabolic traits in Drosophila. Sci. Rep. 2016, 6, 35359. [Google Scholar] [CrossRef] [PubMed]
- Blenau, W.; Baumann, A. Molecular and pharmacological properties of insect biogenic amine receptors: Lessons from Drosophila melanogaster and Apis mellifera. Arch. Insect Biochem. Physiol. 2001, 48, 13–38. [Google Scholar] [CrossRef] [PubMed]
- Blenau, W.; Baumann, A. Aminergic signal transduction in invertebrates: Focus on tyramine and octopamine receptors. Recent Res. Dev. Neurochem. 2003, 6, 225–240. [Google Scholar]
- Evans, P.D.; Maqueira, B. Insect octopamine receptors: A new classification scheme based on studies of cloned Drosophila G-protein coupled receptors. Invert. Neurosci. 2005, 5, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Cazzamali, G.; Williamson, M.; Blenau, W.; Grimmelikhuijzen, C.J. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog. Neurobiol. 2006, 80, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Han, K.A.; Millar, N.S.; Davis, R.L. A novel octopamine receptor with preferential expression in Drosophila mushroom bodies. J. Neurosci. 1998, 18, 3650–3658. [Google Scholar] [PubMed]
- Grohmann, L.; Blenau, W.; Erber, J.; Ebert, P.R.; Strünker, T.; Baumann, A. Molecular and functional characterization of an octopamine receptor from honeybee (Apis mellifera) brain. J. Neurochem. 2003, 86, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Balfanz, S.; Strünker, T.; Frings, S.; Baumann, A. A family of octopamine receptors that specifically induce cyclic AMP production or Ca2+ release in Drosophila melanogaster. J. Neurochem. 2005, 93, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, A.; Arai, Y.; Ozoe, F.; Ohta, H.; Narusuye, K.; Huang, J.; Enomoto, K.; Kataoka, H.; Hirola, A.; Ozoe, Y. Molecular cloning and heterologous expression of an α-adrenergic-like octopamine receptor from the silkworm Bombyx mori. Insect Mol. Biol. 2006, 15, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, H.; Vleugels, R.; Marchal, E.; Badisco, L.; Tobback, J.; Pflüger, H.J.; Blenau, W.; Vanden Broeck, J. The cloning, phylogenetic relationship and distribution pattern of two new putative GPCR-type octopamine receptors in the desert locust (Schistocerca gregaria). J. Insect Physiol. 2010, 56, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Beggs, K.T.; Tyndall, J.D.; Mercer, A.R. Honey bee dopamine and octopamine receptors linked to intracellular calcium signaling have a close phylogenetic and pharmacological relationship. PLoS ONE 2011, 6, e26809. [Google Scholar] [CrossRef] [PubMed]
- Maqueira, B.; Chatwin, H.; Evans, P.D. Identification and characterization of a novel family of Drosophila β-adrenergic-like octopamine G-protein coupled receptors. J. Neurochem. 2005, 94, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ohta, H.; Ozoe, F.; Miyazawa, K.; Huang, J.; Ozoe, Y. Functional and pharmacological characterization of a β-adrenergic-like octopamine receptor from the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2010, 40, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Balfanz, S.; Jordan, N.; Langenstück, T.; Breuer, J.; Bergmeier, V.; Baumann, A. Molecular, pharmacological, and signaling properties of octopamine receptors from honeybee (Apis mellifera) brain. J. Neurochem. 2014, 129, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Saudou, F.; Amlaiky, N.; Plassat, J.L.; Borrelli, E.; Hen, R. Cloning and characterization of a Drosophila tyramine receptor. EMBO J. 1990, 9, 3611–3617. [Google Scholar] [PubMed]
- Vanden Broeck, J.; Vulsteke, V.; Huybrechts, R.; de Loof, A. Characterization of a cloned locust tyramine receptor cDNA by functional expression in permanently transformed Drosophila S2 cells. J. Neurochem. 1995, 64, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Blenau, W.; Balfanz, S.; Baumann, A. Amtyr1: Characterization of a gene from honeybee (Apis mellifera) brain encoding a functional tyramine receptor. J. Neurochem. 2000, 74, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Utsumi, T.; Ozoe, Y. B96Bom encodes a Bombyx mori tyramine receptor negatively coupled to adenylate cyclase. Insect Mol. Biol. 2003, 12, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Utsumi, T.; Ozoe, Y. Amino acid residues involved in interaction with tyramine in the Bombyx mori tyramine receptor. Insect Mol. Biol. 2004, 13, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Mustard, J.A.; Kurshan, P.T.; Hamilton, I.S.; Blenau, W.; Mercer, A.R. Developmental expression of a tyramine receptor gene in the brain of the honey bee, Apis mellifera. J. Comp. Neurol. 2005, 483, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Cazzamali, G.; Klaerke, D.A.; Grimmelikhuijzen, C.J. A new family of insect tyramine receptors. Biochem. Biophys. Res. Commun. 2005, 338, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, A.; Roselli, G.; Evans, P.D. A comparison of the signalling properties of two tyramine receptors from Drosophila. J. Neurochem. 2013, 125, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ohta, H.; Inoue, N.; Takao, H.; Kita, T.; Ozoe, F.; Ozoe, Y. Molecular cloning and pharmacological characterization of a Bombyx mori tyramine receptor selectively coupled to intracellular calcium mobilization. Insect Biochem. Mol. Biol. 2009, 39, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Reim, T.; Balfanz, S.; Baumann, A.; Blenau, W.; Thamm, M.; Scheiner, R. AmTAR2: Functional characterization of a honeybee tyramine receptor stimulating adenylyl cyclase activity. Insect Biochem. Mol. Biol. 2017, 80, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Bischof, L.J.; Enan, E.E. Cloning, expression and functional analysis of an octopamine receptor from Periplaneta americana. Insect Biochem. Mol. Biol. 2004, 34, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Rotte, C.; Krach, C.; Balfanz, S.; Baumann, A.; Walz, B.; Blenau, W. Molecular characterization and localization of the first tyramine receptor of the American cockroach (Periplaneta americana). Neuroscience 2009, 162, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Troppmann, B.; Balfanz, S.; Baumann, A.; Blenau, W. Inverse agonist and neutral antagonist actions of synthetic compounds at an insect 5-HT1 receptor. Br. J. Pharmacol. 2010, 159, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Troppmann, B.; Balfanz, S.; Krach, C.; Baumann, A.; Blenau, W. Characterization of an invertebrate-type dopamine receptor of the American cockroach, Periplaneta americana. Int. J. Mol. Sci. 2014, 15, 629–653. [Google Scholar] [CrossRef] [PubMed]
- Blankenburg, S.; Balfanz, S.; Hayashi, Y.; Shigenobu, S.; Miura, T.; Baumann, O.; Baumann, A.; Blenau, W. Cockroach GABAB receptor subtypes: Molecular characterization, pharmacological properties and tissue distribution. Neuropharmacology 2015, 88, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Blankenburg, S.; Balfanz, S.; Baumann, A.; Blenau, W. Subunit-specific modulatory functions are conserved in an interspecies insect GABAB receptor heteromer. In Advances in Agrochemicals: G Protein-Coupled Receptors (GPCRs) and Ion Channels as Targets for Pest Control, 1st ed.; Coats, J.R., Gross, A.D., Ozoe, Y., Eds.; ACS Publications: Washington, DC, USA, 2017; in press. [Google Scholar]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Merulid, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Hana, S.; Lange, A.B. Cloning and functional characterization of Octβ2-receptor and Tyr1-Receptor in the Chagas disease vector, Rhodnius prolixus. Front. Physiol. 2017, 8, 744. [Google Scholar] [CrossRef] [PubMed]
- Duportets, L.; Barrozo, R.B.; Bozzolan, F.; Gaertner, C.; Anton, S.; Gadenne, C.; Debernard, S. Cloning of an octopamine/tyramine receptor and plasticity of its expression as a function of adult sexual maturation in the male moth Agrotis ipsilon. Insect Mol. Biol. 2010, 19, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Von Nickisch-Rosenegk, E.; Krieger, J.; Kubick, S.; Laage, R.; Strobel, J.; Strotmann, J.; Breer, H. Cloning of biogenic amine receptors from moths (Bombyx mori and Heliothis virescens). Insect Biochem. Mol. Biol. 1996, 26, 817–827. [Google Scholar] [CrossRef]
- Ono, H.; Yoshikawa, H. Identification of amine receptors from a swallowtail butterfly, Papilio xuthus L.: Cloning and mRNA localization in foreleg chemosensory organ for recognition of host plants. Insect Biochem. Mol. Biol. 2004, 34, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Selcho, M.; Pauls, D.; Huser, A.; Stocker, R.F.; Thum, A.S. Characterization of the octopaminergic and tyraminergic neurons in the central brain of Drosophila larvae. J. Comp. Neurol. 2014, 522, 3485–3500. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Rende, E.; Crisanti, A.; Nolan, T. Disruption of aminergic signaling reveals novel compounds with distinct inhibitory effects on mosquito reproduction, locomotor function and survival. Sci. Rep. 2014, 4, 5526. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Stork, T.; Bergles, D.E.; Freeman, M.R. Neuromodulators signal through astrocytes to alter neural circuit activity and behaviour. Nature 2016, 539, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Blumenthal, E.M. Identification of multiple functional receptors for tyramine on an insect secretory epithelium. Sci. Rep. 2017, 7, 168. [Google Scholar] [CrossRef] [PubMed]
- Roeder, T. Tyramine and octopamine: Ruling behavior and metabolism. Annu. Rev. Entomol. 2005, 50, 447–477. [Google Scholar] [CrossRef] [PubMed]
- Kastner, K.W.; Shoue, D.A.; Estiu, G.L.; Wolford, J.; Fuerst, M.F.; Markley, L.D.; Izaguirre, J.A.; McDowell, M.A. Characterization of the Anopheles gambiae octopamine receptor and discovery of potential agonists and antagonists using a combined computational-experimental approach. Malar. J. 2014, 13, 434. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Congreve, M.; Langmead, C.J.; Mason, J.S.; Marshall, F.H. Progress in structure based drug design for G protein-coupled receptors. J. Med. Chem. 2011, 54, 4283–4311. [Google Scholar] [CrossRef] [PubMed]
- Probst, W.C.; Snyder, L.A.; Schuster, D.I.; Brosius, J.; Sealfon, S.C. Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 1992, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bockaert, J.; Pin, J.P. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 1999, 18, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K. G protein selectivity is regulated by multiple intracellular regions of GPCRs. Neurosignals 2003, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rex, E.; Komuniecki, R.W. Characterization of a tyramine receptor from Caenorhabditis elegans. J. Neurochem. 2002, 82, 1352–1359. [Google Scholar] [CrossRef] [PubMed]
- Rex, E.; Molitor, S.C.; Hapiak, V.; Xiao, H.; Henderson, M.; Komuniecki, R. Tyramine receptor (SER-2) isoforms are involved in the regulation of pharyngeal pumping and foraging behavior in Caenorhabditis elegans. J. Neurochem. 2004, 91, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Uzzan, A.; Dudai, Y. Aminergic receptors in Drosophila melanogaster: Responsiveness of adenylate cyclase to putative neurotransmitters. J. Neurochem. 1982, 38, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, M.; Nakane, T.; Ono, T.; Khan, M.A.; Ohta, H.; Ozoe, Y. Substituent-dependent, positive and negative modulation of Bombyx mori adenylate cyclase by synthetic octopamine/tyramine analogues. Arch. Insect Biochem. Physiol. 2001, 47, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Nakane, T.; Ohta, H.; Ozoe, Y. Positive and negative modulation of Bombyx mori adenylate cyclase by 5-phenyloxazoles: Identification of octopamine and tyramine receptor agonists. Arch. Insect Biochem. Physiol. 2003, 52, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Blenau, W.; May, T.; Erber, J. Characterization of [3H]LSD binding to a serotonin-sensitive site in honeybee (Apis mellifera) brain. Comp. Biochem. Physiol. B 1995, 112, 377–384. [Google Scholar] [CrossRef]
- Röser, C.; Jordan, N.; Balfanz, S.; Baumann, A.; Walz, B.; Baumann, O.; Blenau, W. Molecular and pharmacological characterization of serotonin 5-HT2α and 5-HT7 receptors in the salivary glands of the blowfly Calliphora vicina. PLoS ONE 2012, 7, e49459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thamm, M.; Rolke, D.; Jordan, N.; Balfanz, S.; Schiffer, C.; Baumann, A.; Blenau, W. Function and distribution of 5-HT2 receptors in the honeybee (Apis mellifera). PLoS ONE 2013, 8, e82407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blenau, W.; Stöppler, D.; Balfanz, S.; Thamm, M.; Baumann, A. Dm5-HT2B: Pharmacological characterization of the fifth serotonin receptor subtype of Drosophila melanogaster. Front. Syst. Neurosci. 2017, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Minhas, N.; Gole, J.W.D.; Orr, G.L.; Downer, R.G.H. Pharmacology of [3H]mianserin binding in the nerve cord of the American cockroach, Periplaneta americana. Arch. Insect Biochem. Physiol. 1987, 6, 191–201. [Google Scholar] [CrossRef]
- Roeder, T. High-affinity antagonists of the locust neuronal octopamine receptor. Eur. J. Pharmacol. 1990, 191, 221–224. [Google Scholar] [CrossRef]
- Blenau, W.; Baumann, A. Molecular characterization of the ebony gene from the American cockroach, Periplaneta americana. Arch. Insect Biochem. Physiol. 2005, 59, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kozak, M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984, 12, 857–872. [Google Scholar] [CrossRef] [PubMed]
- Thamm, M.; Balfanz, S.; Scheiner, R.; Baumann, A.; Blenau, W. Characterization of the 5-HT1A receptor of the honeybee (Apis mellifera) and involvement of serotonin in phototactic behavior. Cell. Mol. Life Sci. 2010, 67, 2467–2479. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, J.; Margalit, T.; Eismann, E.; Lancet, D.; Kaupp, U.B. Primary structure of cAMP-gated channel from bovine olfactory epithelium. FEBS Lett. 1990, 270, 24–29. [Google Scholar] [CrossRef]
- Chen, C.; Okayama, H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987, 7, 2745–2752. [Google Scholar] [CrossRef] [PubMed]
- Wachten, S.; Schlenstedt, J.; Gauss, R.; Baumann, A. Molecular identification and functional characterization of an adenylyl cyclase from the honeybee. J. Neurochem. 2006, 96, 1580–1590. [Google Scholar] [CrossRef] [PubMed]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blenau, W.; Balfanz, S.; Baumann, A. PeaTAR1B: Characterization of a Second Type 1 Tyramine Receptor of the American Cockroach, Periplaneta americana. Int. J. Mol. Sci. 2017, 18, 2279. https://doi.org/10.3390/ijms18112279
Blenau W, Balfanz S, Baumann A. PeaTAR1B: Characterization of a Second Type 1 Tyramine Receptor of the American Cockroach, Periplaneta americana. International Journal of Molecular Sciences. 2017; 18(11):2279. https://doi.org/10.3390/ijms18112279
Chicago/Turabian StyleBlenau, Wolfgang, Sabine Balfanz, and Arnd Baumann. 2017. "PeaTAR1B: Characterization of a Second Type 1 Tyramine Receptor of the American Cockroach, Periplaneta americana" International Journal of Molecular Sciences 18, no. 11: 2279. https://doi.org/10.3390/ijms18112279
APA StyleBlenau, W., Balfanz, S., & Baumann, A. (2017). PeaTAR1B: Characterization of a Second Type 1 Tyramine Receptor of the American Cockroach, Periplaneta americana. International Journal of Molecular Sciences, 18(11), 2279. https://doi.org/10.3390/ijms18112279